PERFF-seq
Transcript-specific enrichment enables profiling of rare cell states via single-cell RNA sequencing
Single-cell genomics technologies have accelerated our understanding of cell-state heterogeneity in nearly every complex tissue. While single-cell genomics can identify rare populations that express specific marker transcript combinations, traditional flow sorting requires cell surface markers with high-fidelity antibodies, limiting followup study, particularly when nuclei isolation is required. Programmable Enrichment via RNA Flow-FISH by sequencing (PERFF-seq) is our solution to these challenges by enabling scRNA-seq profiling of subpopulations defined by the abundance of specific RNA transcripts.
Here’s a hub for the key links associated with PERFF-seq.
Critical reagents
- Molecular Instruments HCR-FlowFISH
- 10x Genomics Flex scRNA-seq
- Thermo Fisher Scientific dsDNase
Key links
- Go behind the paper with a Q&A from Tsion!
- Find the full work available at Nature Genetics
- Read the step-by-step protocols.io workflow for PERFF-seq and cell/nuclei processing
- Reproduce all custom code and analyses from our Github Repository
- Access all raw and processed data available on GEO
- Find an easy-to-read digest of our results here
- Easily prepare buffers and reagents with the PERFF-seq benchside notes
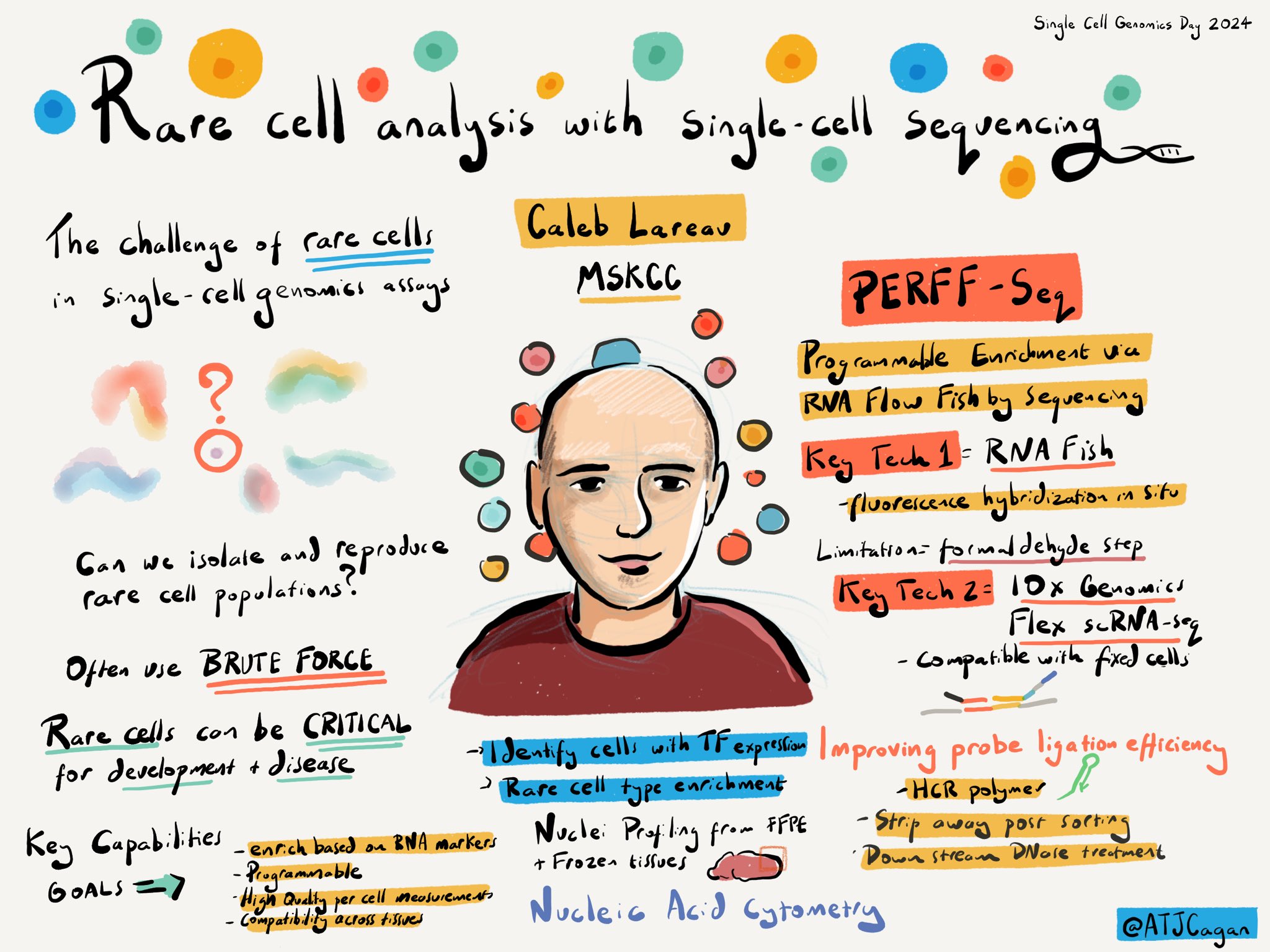
Live sketch summary of Caleb’s talk at scgd24 by Alex Cagan
Contact the authors