What are barcode multiplets?
A common / foundational assumption in single-cell genomics analyses is that a single oligonucleotide barcode represents the nucleic acid of an individual cell. However, there are scenarios where an individual single cell can have its contents barcoded by multiple distinct oligonucleotides, termed barcode multiplets. These scenarios include i) a single droplet containing multiple beads or ii) multiple oligonucleotides present on the same individual bead. Every single-cell assay has barcode multiplets to varying degrees.
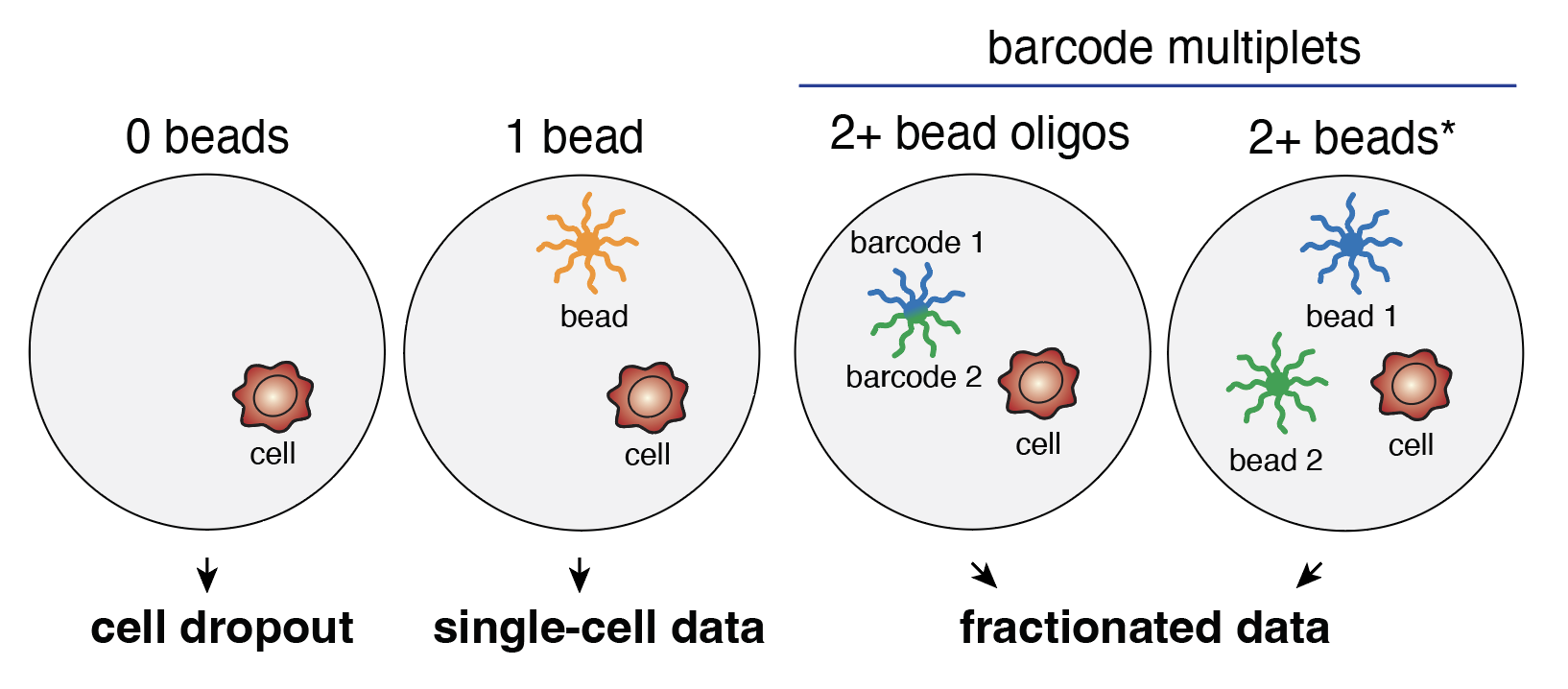
Barcode multiplets used to develop dscATAC-seq
Prior work has shown that if you adjust the size/composition of the beads, one can control the number of beads loaded into each droplet. 10x Genomics uses these soft hydrogel beads to ensure that you get 1 bead per droplet.
For small beads, including those used in Drop-seq and other home-brew settings, this is hard to achieve, including in the commercialized BioRad ddSeq platform. This is an obstacle because you split data
To get around this, we superloaded beads to reduce the number of droplets with zero beads. In the process, we greatly increase the number of droplets with 2+ bead Fortunately, due to droplet-PCR, beads in the same droplet will share the same transposition positions of genomic fragments, enabling a statistic that can pair barcode multiplets. We call this tool bap that can detect barcode multiplets efficently.
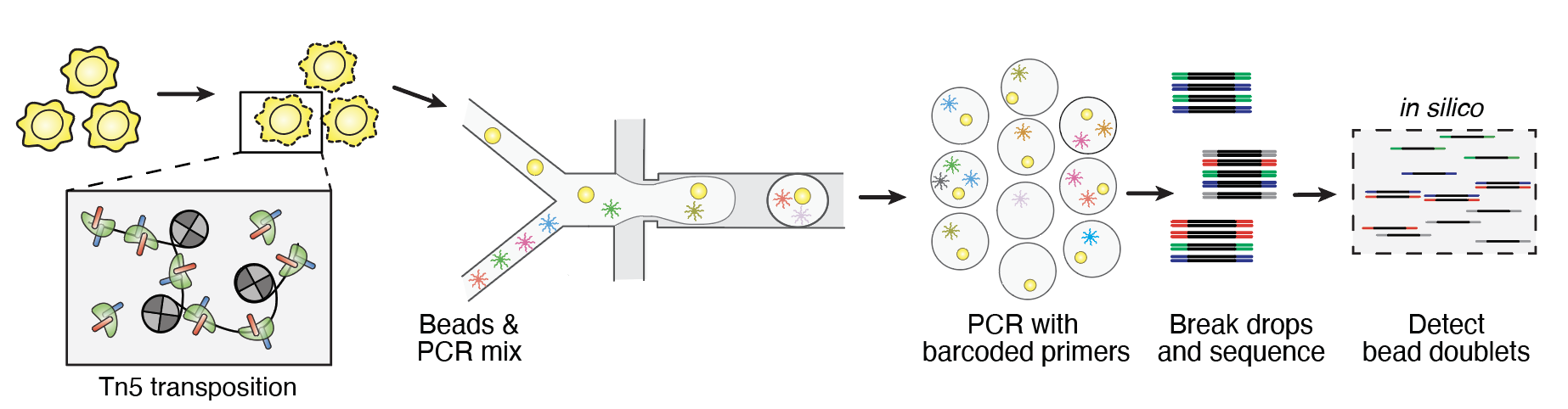
Using bap, the BioRad ddSEQ ATAC
kit could become a reality. By merging barcode multiplets, the approach had very low cell dropout / high cell recovery.
Accuracy of bap
Implicit in all of this is the requirement of bap to be very accurate– you wanted to merge everything
that should be merged and miss nothing that shouldn’t. To test this, we designed an experiment with
diverse exogenous oligonucleotides to establish a set of true beads that were co-contained in droplets
alongside capture of a cell and then ran bap to recover the pairs of barcodes.
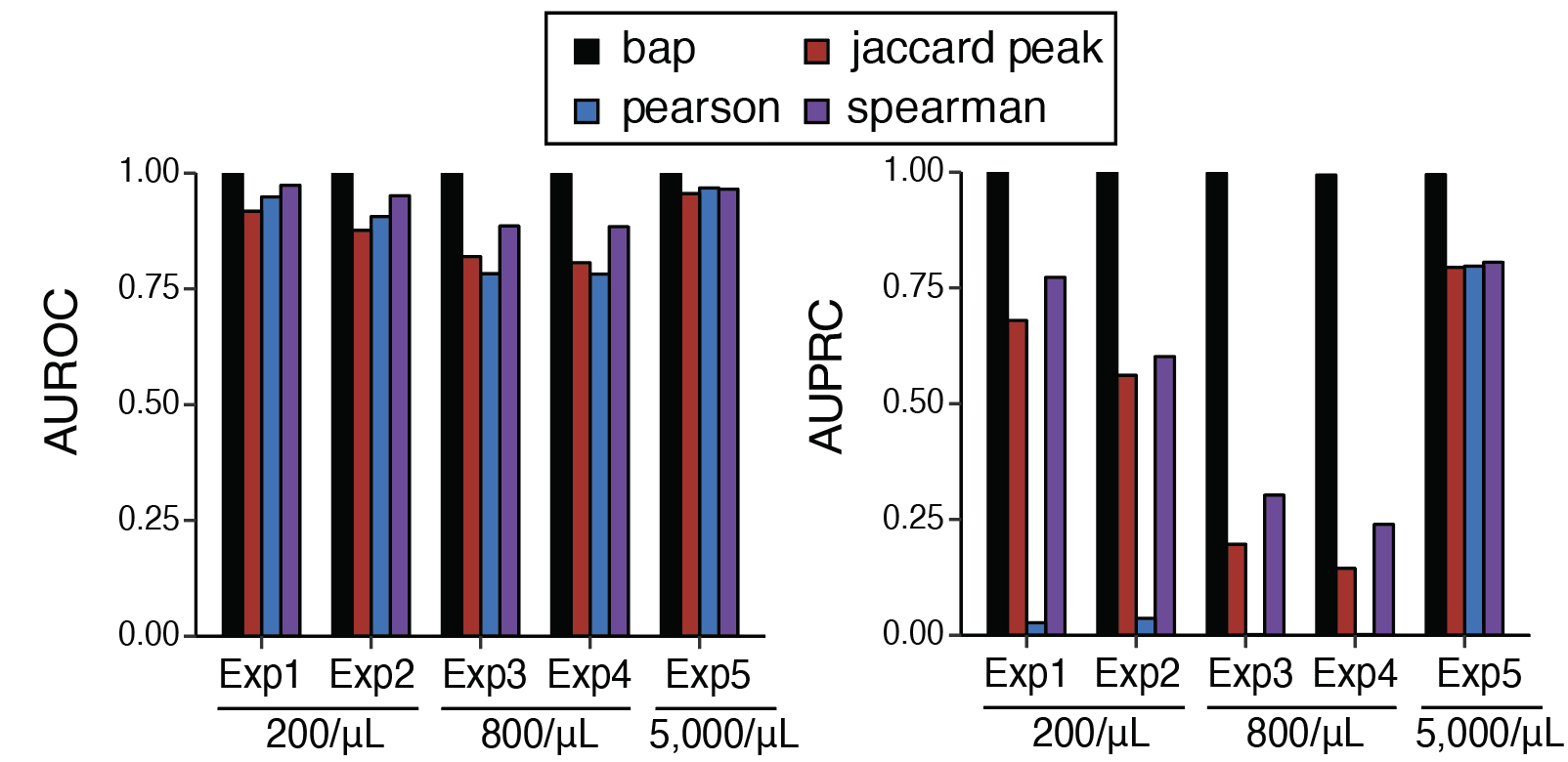
The results were clear: at any bead concentration, bap had an AUROC/AUPRC of 0.999+,
meaning we could trivially classify barcode multiplets from single-cell data.
Application to 10x Genomics scATAC data
When applying bap to scATAC-seq datasets released from 10x Genomics, instances of 9+ bead barcodes were annotated by bap of belonging to the same droplet (all barcodes annotated as ‘cells’ having a sequence in the whitelist and read abundances above the knee threshold). These barcodes were enriched in the same biological cluster, supportive of the idea that they may be ‘replicates’ of the same cell.
fully supports processing 10x Genomics data.
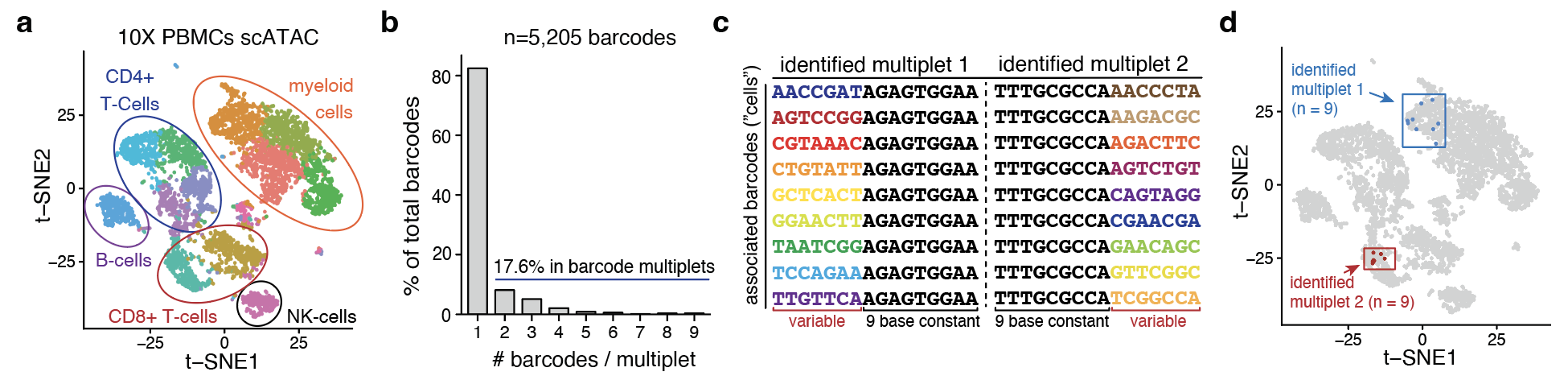
Taking a closer look at the underlying barcode sequence, a common 7 nucleotide variable region followed by a 9 base constant in these barcodes, consistent with the idea that multiple ‘whitelisted’ barcode sequences were installed on an individual bead during synthesis.
As far as we know, this issue was prevalent in the early ATAC beads that were synthesized by 10x but have been since resolved in the ATAC and other kits. Further, a software implementation (that gives nearly identical results to bap) realeased from 10x Genomics was incorporated into CellRanger-ATAC v1.2. This functionality similarly analyzes shared transposition events between pairs of barcode sequences to identify multiplets. Whereas bap merges different bead barcode sequences into a single cell barcode, the implementation in CellRanger-ATAC simply discards multiplet barcodes after retaining the one bead barcode with the highest number of fragments.
In short, as long as you are using v1.2+ of CellRanger-ATAC, you shouldn’t have to worry about barcode multiplets.