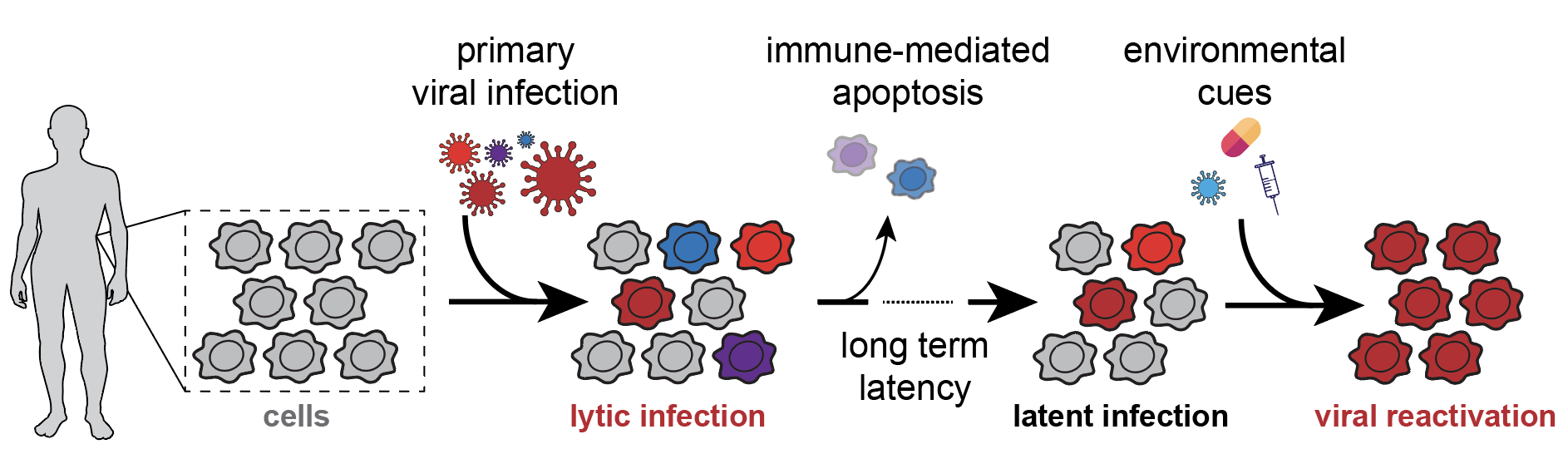
Overview of detection methods
Repo for the main code associated with this work is available here
Use Serratus to perform a human
For a digest on how to screen all human viruses by all RNA-seq libraries in Serratus, check out the repo here for reproducing the pan-virus screen.
HHV-6B reference genome
We developed a kallisto|bustools workflow to rapidly quantify reads from either single-cell or bulk transcriptomes.
We downloaded the GenBank AF157706 reference transcriptome and created a kallisto index using the default -k 31 (kmer) parameter.
The best place for reproducing our HHV-6 pseudoalignment pipeline is here.
For single-cell libraries, raw sequencing reads were processed using the kallisto bus command with appropriate hyperparameters for each version
of the single-cell chemistry (either 14 or 16 bp sequence barcode and 10 or 12 bases of UMI sequence).
After barcode and UMI correction, a plain text sparse matrix was emitted, corresponding to unique HHV-6B reads
mapping to individual cells in the single-cell sequencing library. For bulk libraries, the same index could be utilized with the standard kallisto quant execution.