Publications
2026

Population-scale sequencing resolves determinants of persistent EBV DNA
Nature
·
28 Jan 2026
·
doi:10.1038/s41586-025-10020-2

Characterization of a pathogenic subpopulation of human glioma associated macrophages linked to glioma progression
Cancer Cell
·
01 Jan 2026
·
doi:10.1016/j.ccell.2025.12.010
2025
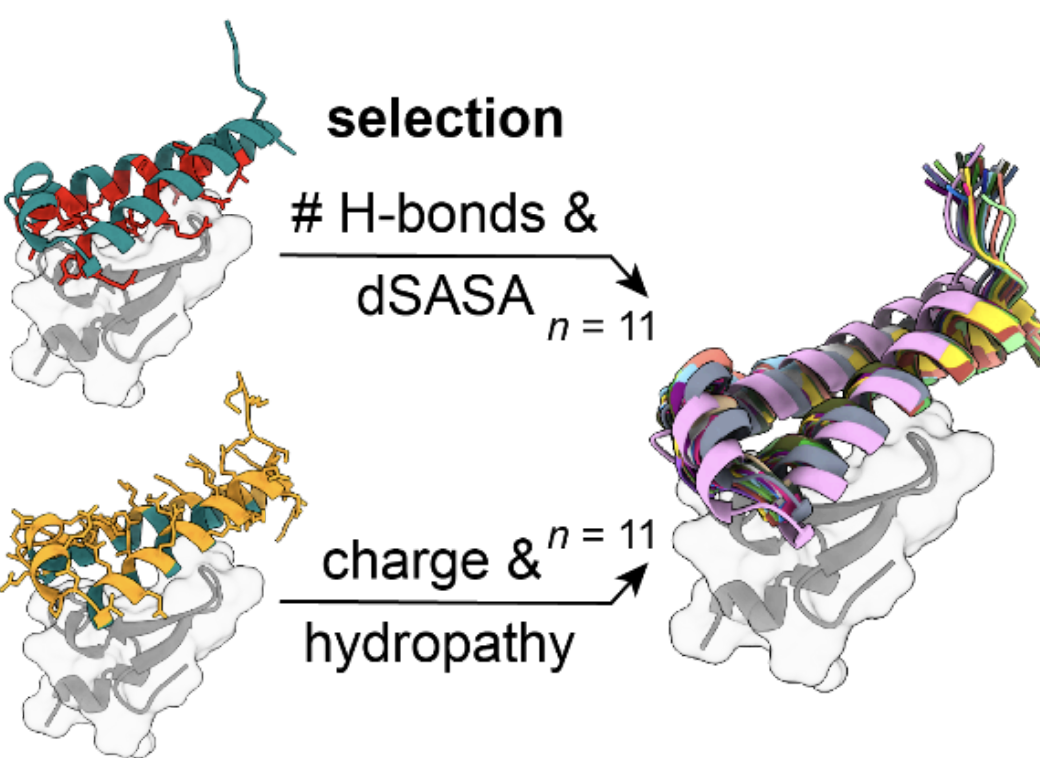
Sequence and structural determinants of efficacious
de novo
chimeric antigen receptors
openRxiv
·
13 Dec 2025
·
doi:10.64898/2025.12.12.694033
Independent mechanisms of inflammation and myeloid bias in VEXAS syndrome
Nature
·
03 Nov 2025
·
doi:10.1038/s41586-025-09815-0
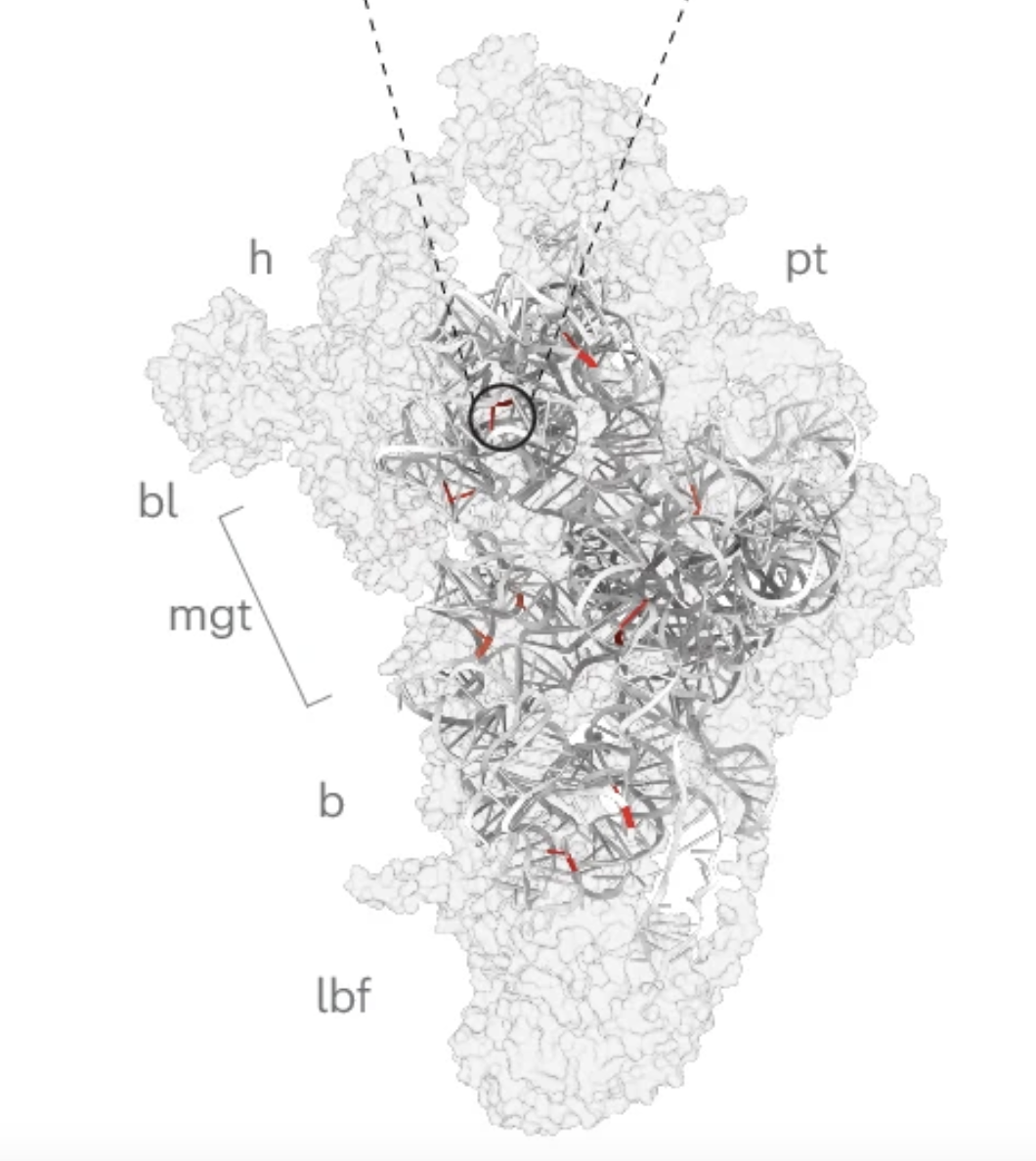
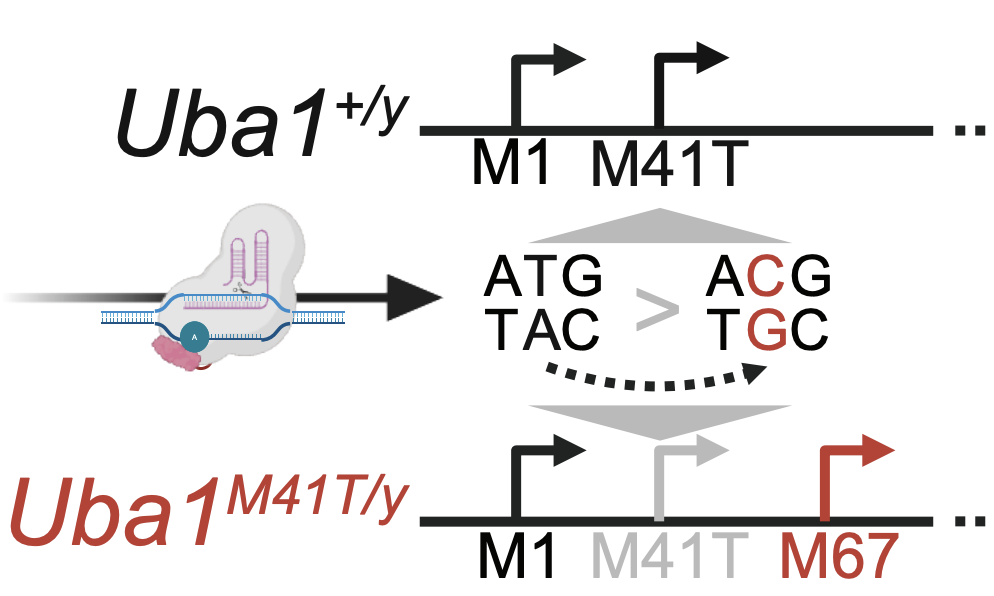
Functionally dominant hotspot mutations of mitochondrial ribosomal RNA genes in cancer
Nature Genetics
·
01 Nov 2025
·
doi:10.1038/s41588-025-02374-0
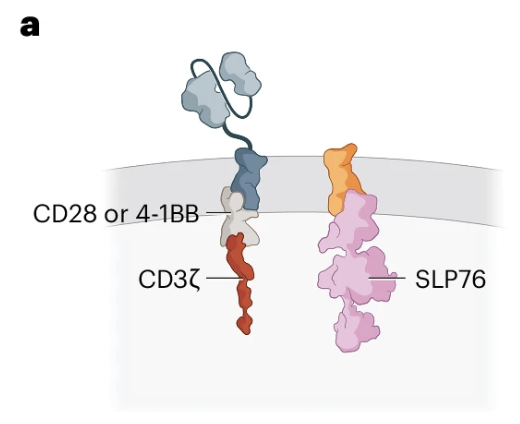
Engineering T cells with a membrane-tethered version of SLP-76 overcomes antigen-low resistance to CAR T cell therapy
Nature Cancer
·
23 Oct 2025
·
doi:10.1038/s43018-025-01056-4
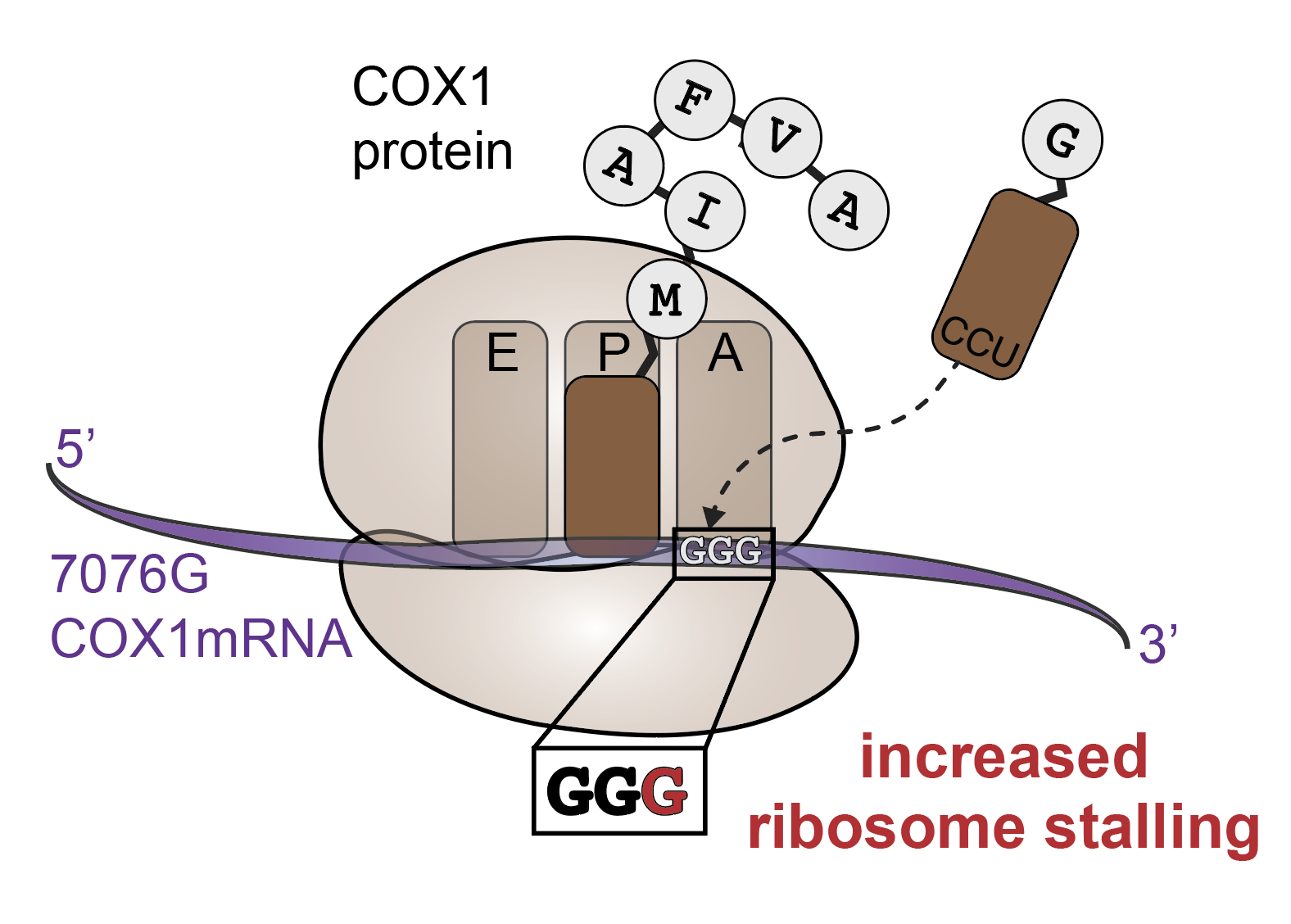
Cell type–specific purifying selection of synonymous mitochondrial DNA variation
Proceedings of the National Academy of Sciences
·
24 Jul 2025
·
doi:10.1073/pnas.2505704122

CAR-engineered lymphocyte persistence is governed by a FAS ligand–FAS autoregulatory circuit
Nature Cancer
·
22 Jul 2025
·
doi:10.1038/s43018-025-01009-x

Clonal lineage tracing of innate immune cells in human cancer
Cold Spring Harbor Laboratory
·
21 Jul 2025
·
doi:10.1101/2025.07.16.665245

Methods for multiplexing single-cell multi-omics
Nature Methods
·
26 May 2025
·
doi:10.1038/s41592-025-02657-8

Clonal tracing with somatic epimutations reveals dynamics of blood ageing
Nature
·
21 May 2025
·
doi:10.1038/s41586-025-09041-8

Single-cell multi-omics resolved analysis of mitochondrial genome-wide mutational burden, constraint, and mosaicism
Springer Science and Business Media LLC
·
21 May 2025
·
doi:10.21203/rs.3.rs-6539733/v1

Engineering mtDNA deletions by reconstituting end joining in human mitochondria
Cell
·
01 May 2025
·
doi:10.1016/j.cell.2025.02.009
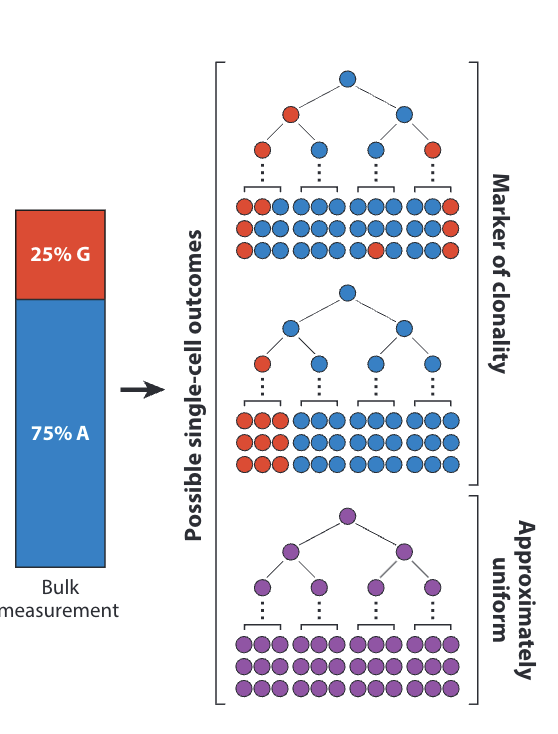
Single-Cell Technologies for Studying the Evolution and Function of Mitochondrial DNA Heteroplasmy in Cancer
Annual Review of Cancer Biology
·
11 Apr 2025
·
doi:10.1146/annurev-cancerbio-080124-102241
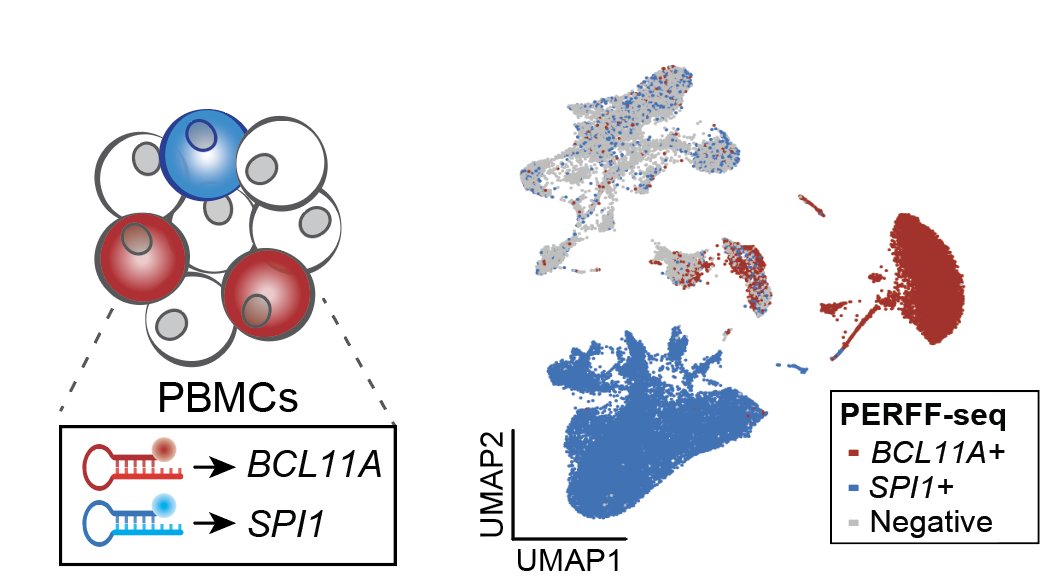
Transcript-specific enrichment enables profiling of rare cell states via single-cell RNA sequencing
Nature Genetics
·
08 Jan 2025
·
doi:10.1038/s41588-024-02036-7
2024

Dynamics of Immune Reconstitution and Impact on Outcomes across CAR-T Cell Products in Large B-cell Lymphoma
Blood Cancer Discovery
·
12 Dec 2024
·
doi:10.1158/2643-3230.BCD-24-0163
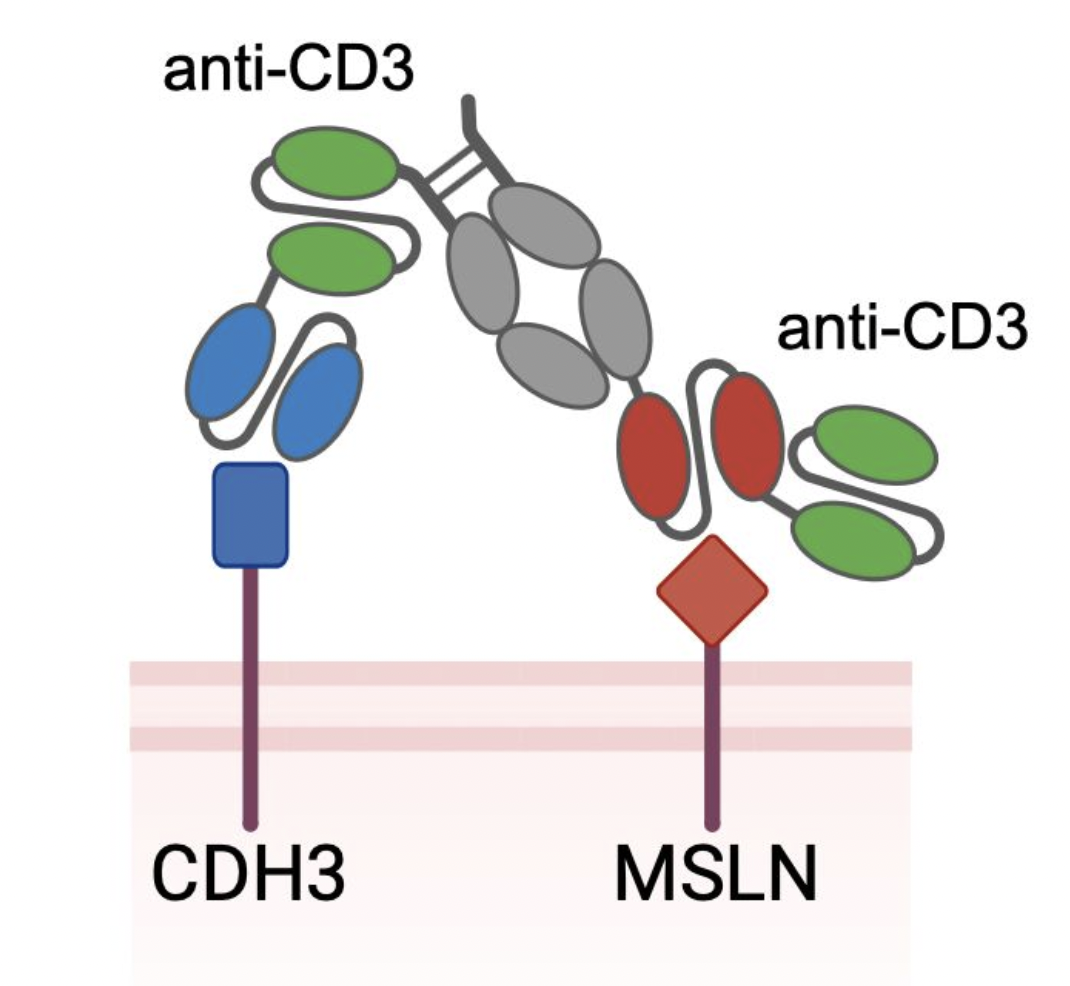
Identifying optimal tumor-associated antigen combinations with single-cell genomics to enable multi-targeting therapies
Frontiers in Immunology
·
07 Nov 2024
·
doi:10.3389/fimmu.2024.1492782
Retinoic acid and TGF-β orchestrate organ-specific programs of tissue residency
Immunity
·
01 Nov 2024
·
doi:10.1016/j.immuni.2024.09.015
Concepts and new developments in droplet-based single cell multi-omics
Trends in Biotechnology
·
01 Nov 2024
·
doi:10.1016/j.tibtech.2024.07.006
Tracking Rare Single Donor and Recipient Immune and Leukemia Cells after Allogeneic Hematopoietic Cell Transplantation Using Mitochondrial DNA Mutations
Blood Cancer Discovery
·
05 Sep 2024
·
doi:10.1158/2643-3230.BCD-23-0138
Deciphering the impact of genomic variation on function
Nature
·
04 Sep 2024
·
doi:10.1038/s41586-024-07510-0
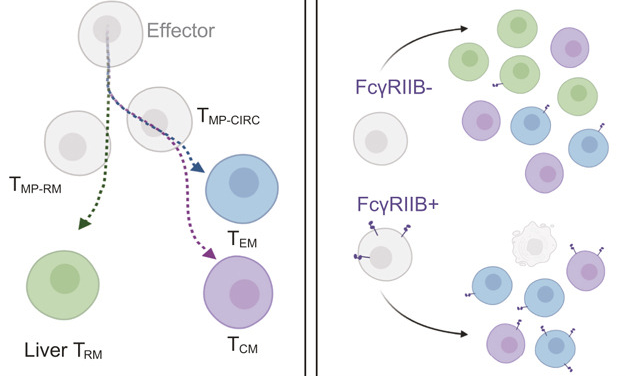
Distinct epigenomic landscapes underlie tissue-specific memory T cell differentiation
Immunity
·
01 Sep 2024
·
doi:10.1016/j.immuni.2024.06.014
Logan: Planetary-Scale Genome Assembly Surveys Life’s Diversity
Cold Spring Harbor Laboratory
·
31 Jul 2024
·
doi:10.1101/2024.07.30.605881
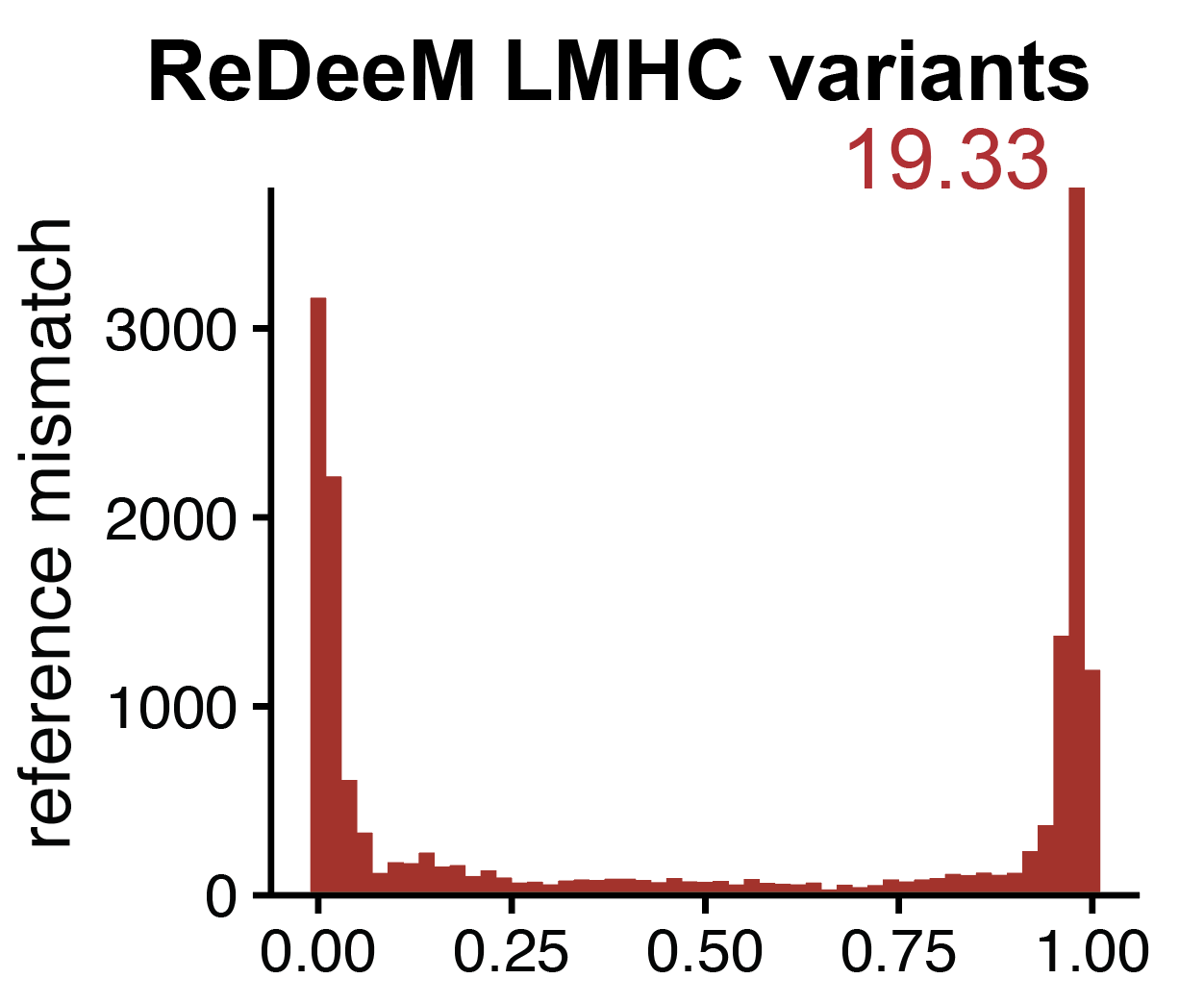
Artifacts in single-cell mitochondrial DNA mutation analyses misinform phylogenetic inference
Cold Spring Harbor Laboratory
·
29 Jul 2024
·
doi:10.1101/2024.07.28.605517
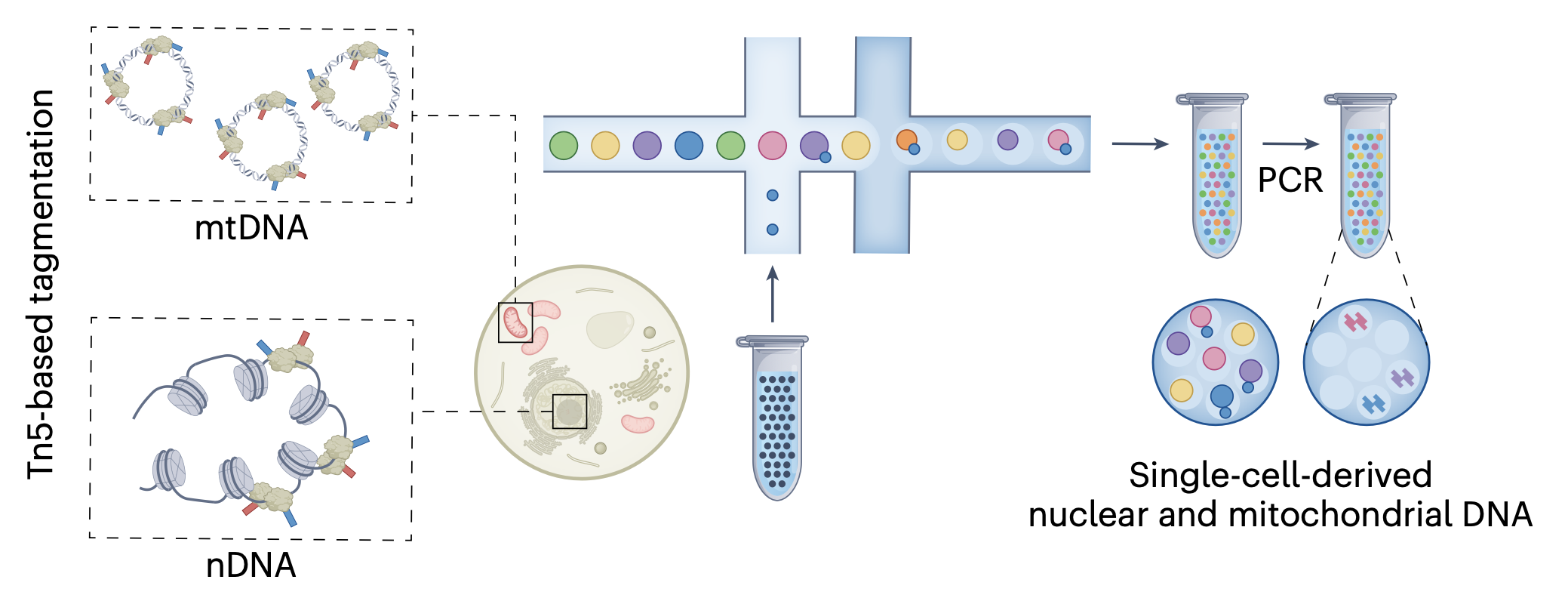
Mitochondrial genetics through the lens of single-cell multi-omics
Nature Genetics
·
01 Jul 2024
·
doi:10.1038/s41588-024-01794-8
Functional dissection of complex and molecular trait variants at single nucleotide resolution
Cold Spring Harbor Laboratory
·
06 May 2024
·
doi:10.1101/2024.05.05.592437
A CTCF-binding site in the Mdm1-Il22-Ifng locus shapes cytokine expression profiles and plays a critical role in early Th1 cell fate specification
Immunity
·
01 May 2024
·
doi:10.1016/j.immuni.2024.04.007
Convergent epigenetic evolution drives relapse in acute myeloid leukemia
eLife
·
22 Apr 2024
·
doi:10.7554/eLife.93019
FOXO1 is a master regulator of memory programming in CAR T cells
Nature
·
10 Apr 2024
·
doi:10.1038/s41586-024-07300-8
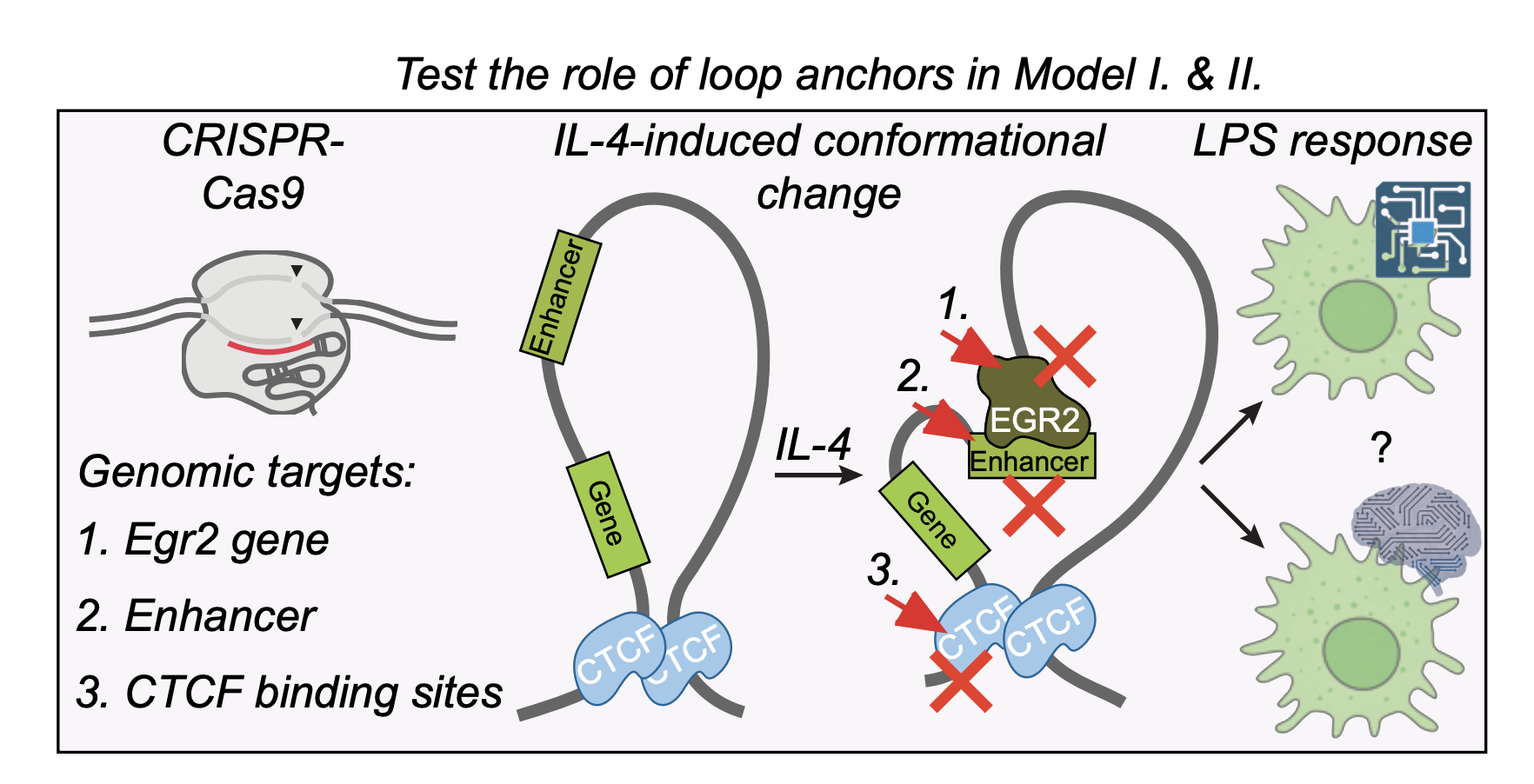
Regulation of immune signal integration and memory by inflammation-induced chromosome conformation
Cold Spring Harbor Laboratory
·
04 Mar 2024
·
doi:10.1101/2024.02.29.582872
2023
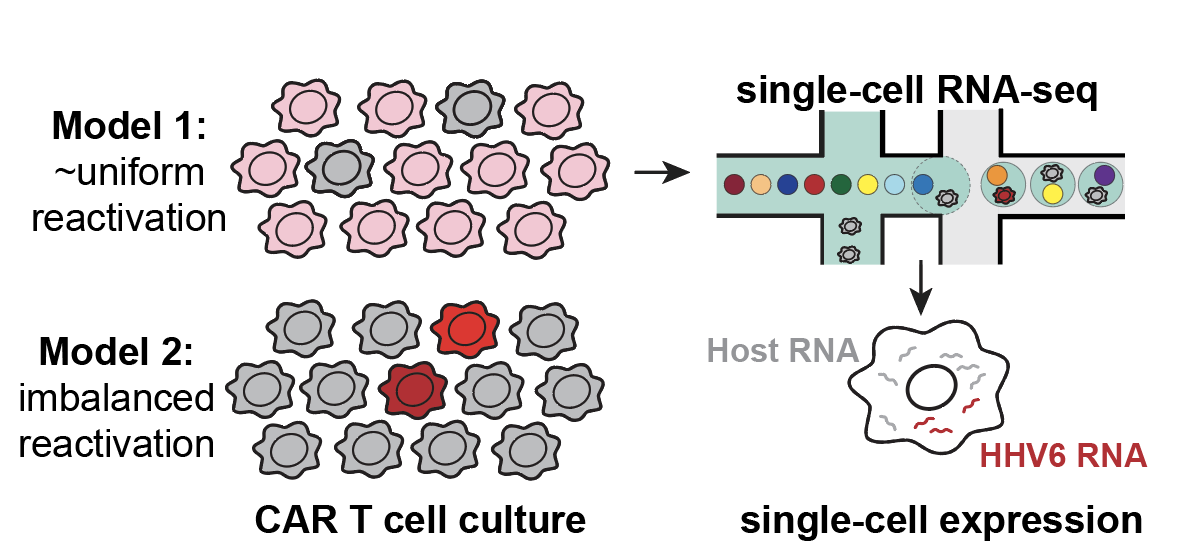
Latent human herpesvirus 6 is reactivated in CAR T cells
Nature
·
08 Nov 2023
·
doi:10.1038/s41586-023-06704-2

Multi-modal skin atlas identifies a multicellular immune-stromal community associated with altered cornification and specific T cell expansion in atopic dermatitis
Cold Spring Harbor Laboratory
·
30 Oct 2023
·
doi:10.1101/2023.10.29.563503
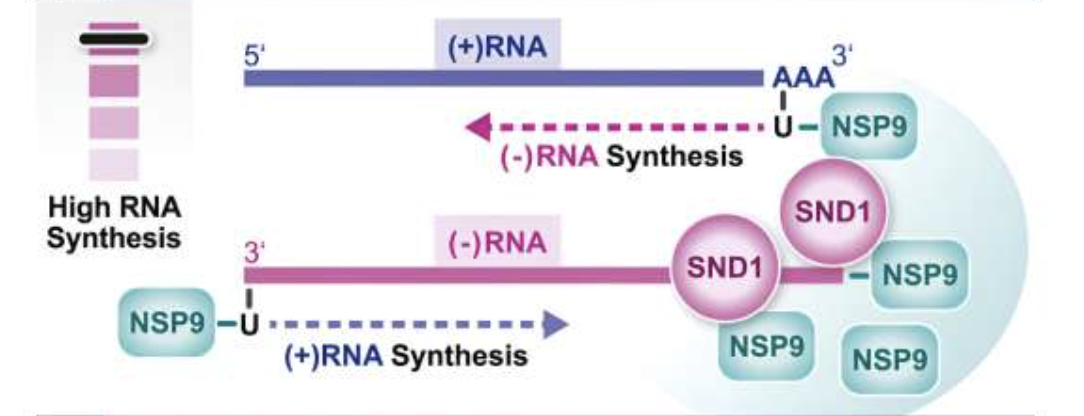
SND1 binds SARS-CoV-2 negative-sense RNA and promotes viral RNA synthesis through NSP9
Cell
·
01 Oct 2023
·
doi:10.1016/j.cell.2023.09.002
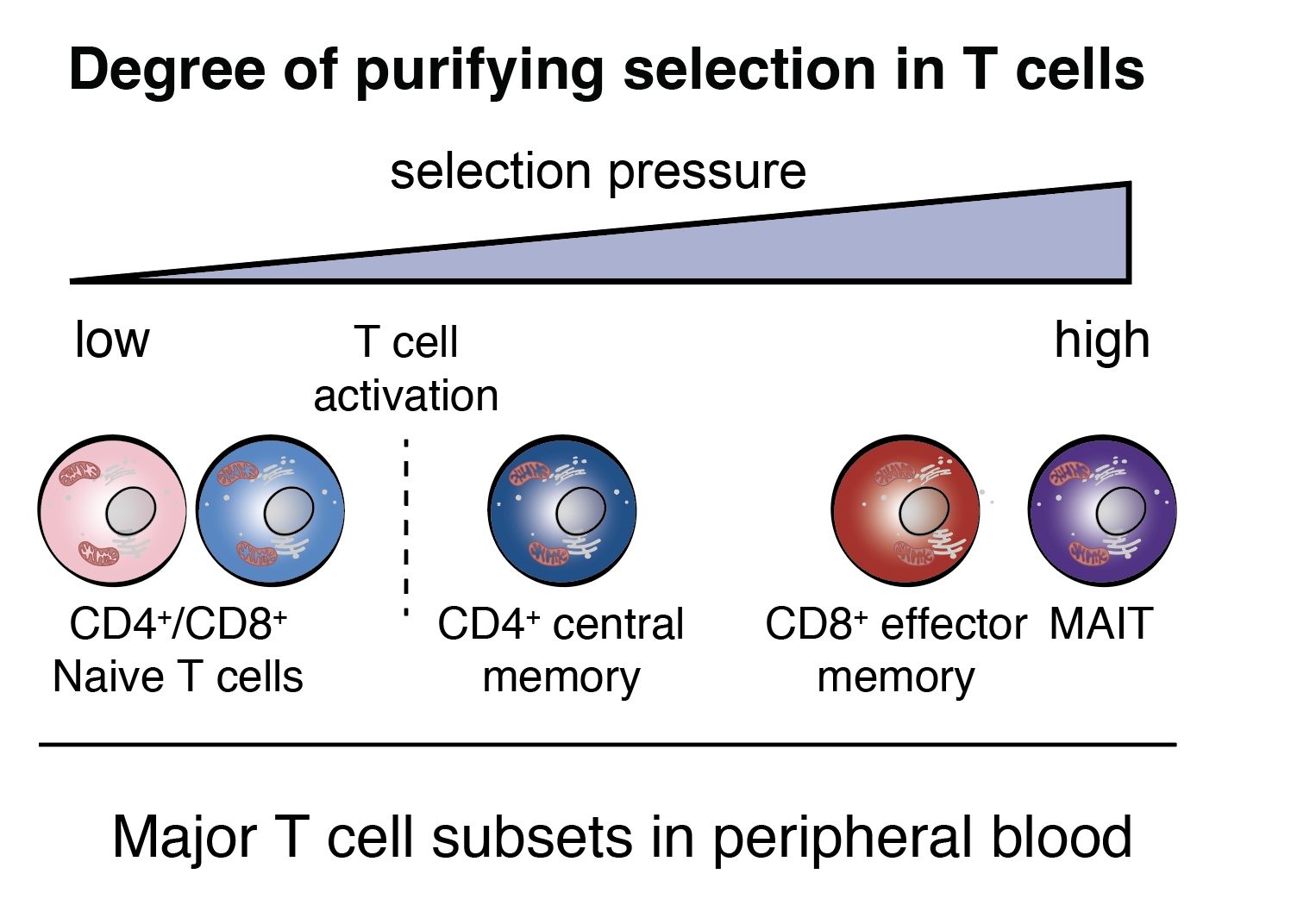
Single-cell multi-omics of mitochondrial DNA disorders reveals dynamics of purifying selection across human immune cells
Nature Genetics
·
29 Jun 2023
·
doi:10.1038/s41588-023-01433-8
Subtle cell states resolved in single-cell data
Nature Biotechnology
·
17 May 2023
·
doi:10.1038/s41587-023-01797-6
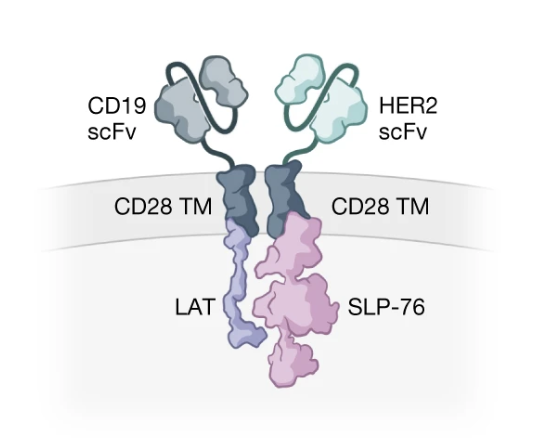
Co-opting signalling molecules enables logic-gated control of CAR T cells
Nature
·
08 Mar 2023
·
doi:10.1038/s41586-023-05778-2
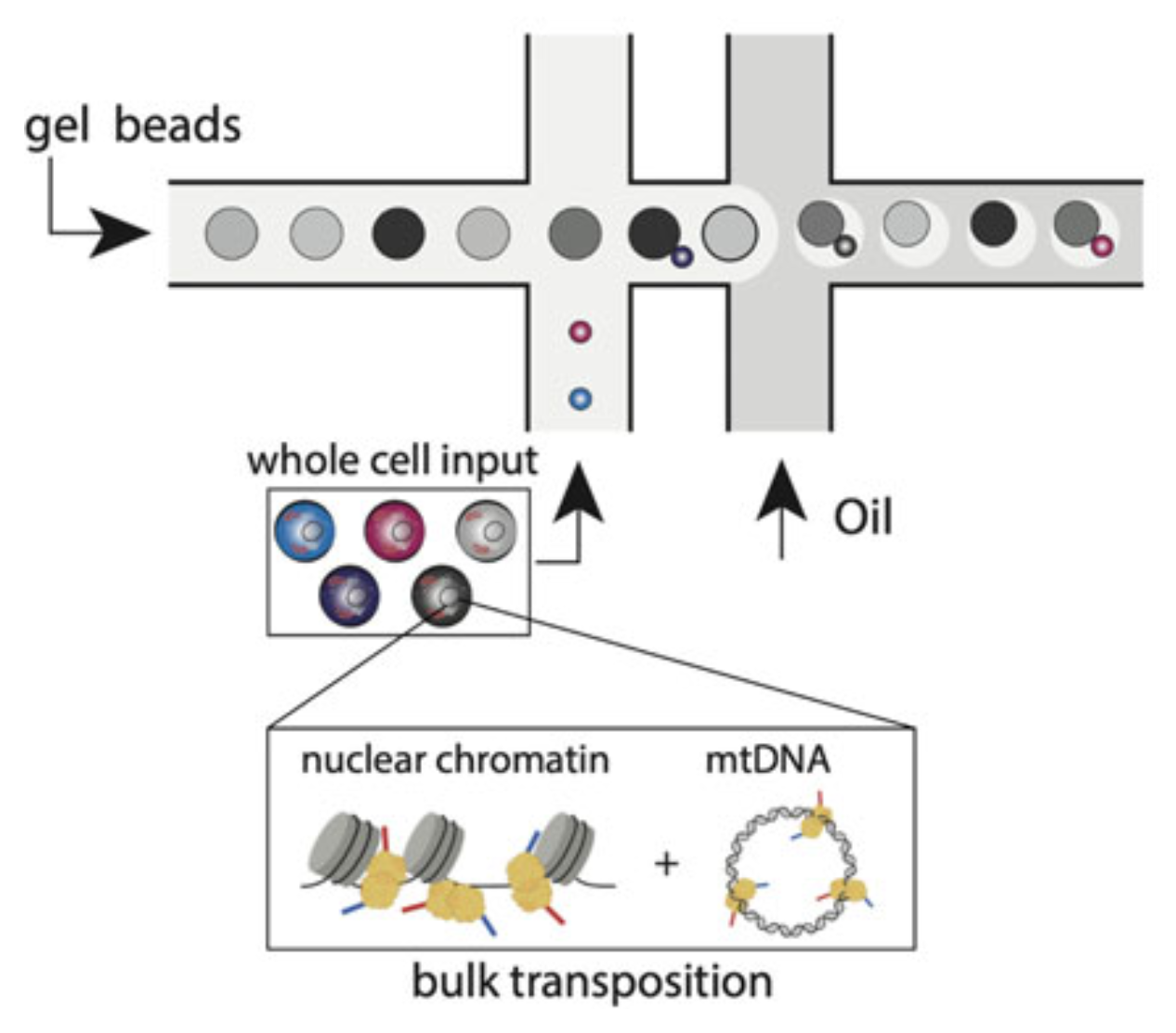
Mitochondrial single-cell ATAC-seq for high-throughput multi-omic detection of mitochondrial genotypes and chromatin accessibility
Nature Protocols
·
15 Feb 2023
·
doi:10.1038/s41596-022-00795-3
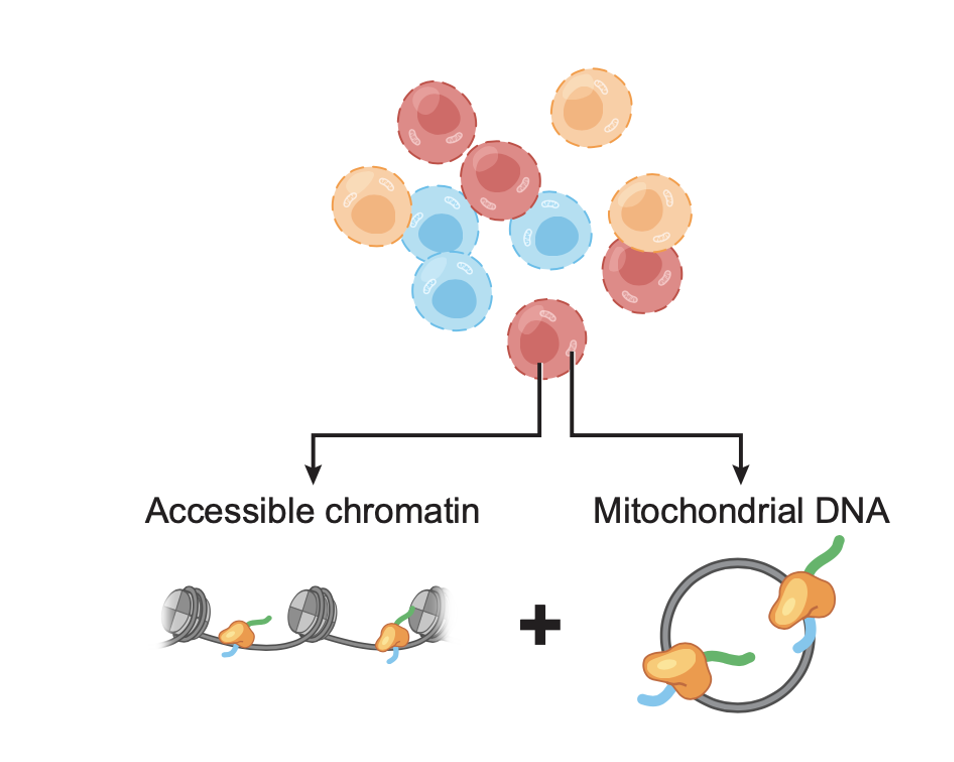
Concomitant Sequencing of Accessible Chromatin and Mitochondrial Genomes in Single Cells Using mtscATAC-Seq
Methods in Molecular Biology
·
01 Jan 2023
·
doi:10.1007/978-1-0716-2899-7_14
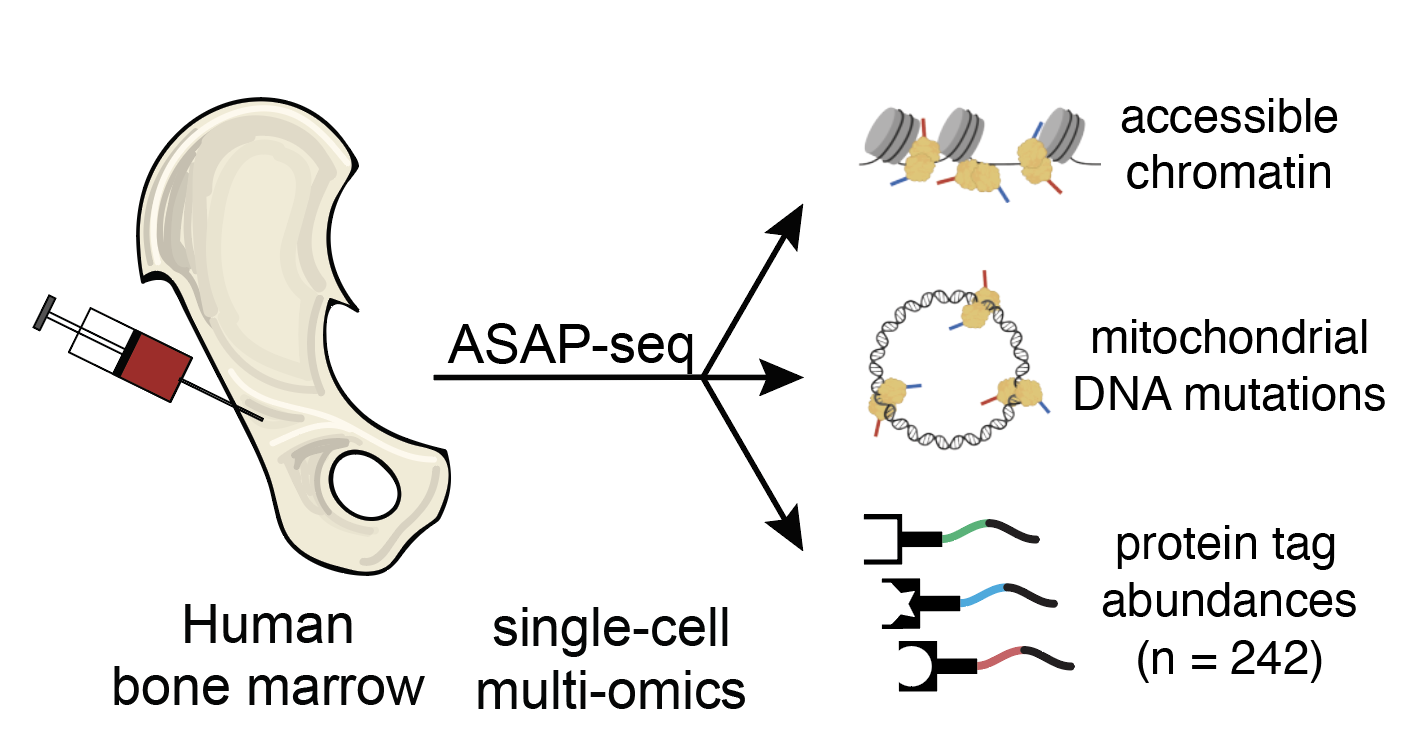
Massively Parallel Profiling of Accessible Chromatin and Proteins with ASAP-Seq
Methods in Molecular Biology
·
01 Jan 2023
·
doi:10.1007/978-1-0716-2899-7_13
2022
Mitochondrial DNA Mutations as Natural Barcodes for Lineage Tracing of Murine Tumor Models
Cancer Research
·
05 Dec 2022
·
doi:10.1158/0008-5472.CAN-22-0275
BAF Complex Maintains Glioma Stem Cells in Pediatric H3K27M Glioma
Cancer Discovery
·
28 Oct 2022
·
doi:10.1158/2159-8290.CD-21-1491
Clonal expansion and epigenetic inheritance of long-lasting NK cell memory
Nature Immunology
·
26 Oct 2022
·
doi:10.1038/s41590-022-01327-7

Editorial: Lineage tracing, hematopoietic stem cell and immune cell dynamics
Frontiers in Immunology
·
18 Oct 2022
·
doi:10.3389/fimmu.2022.1062415
A RORγt+ cell instructs gut microbiota-specific Treg cell differentiation
Nature
·
07 Sep 2022
·
doi:10.1038/s41586-022-05089-y
Functional inference of gene regulation using single-cell multi-omics
Cell Genomics
·
01 Sep 2022
·
doi:10.1016/j.xgen.2022.100166
Advancing T cell–based cancer therapy with single-cell technologies
Nature Medicine
·
01 Sep 2022
·
doi:10.1038/s41591-022-01986-x
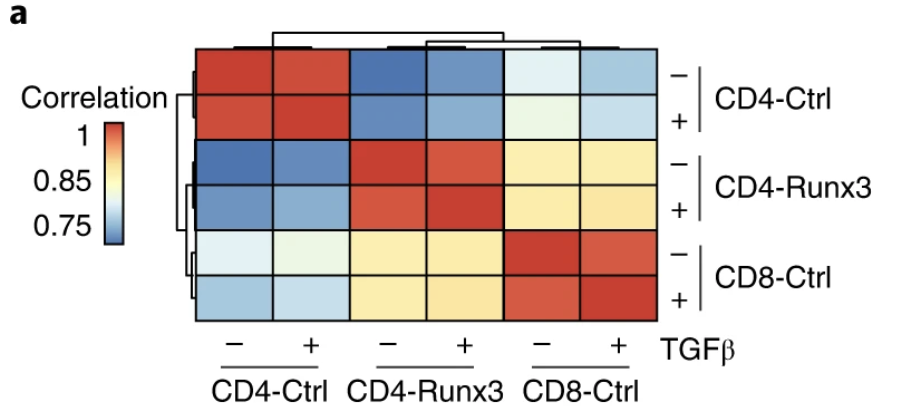
Runx3 drives a CD8+ T cell tissue residency program that is absent in CD4+ T cells
Nature Immunology
·
26 Jul 2022
·
doi:10.1038/s41590-022-01273-4
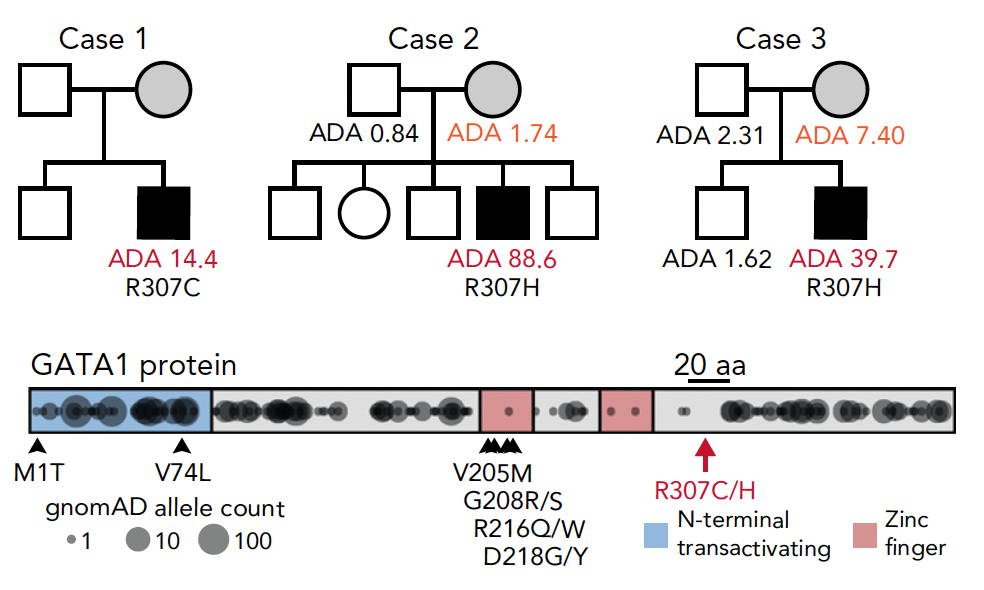
Congenital anemia reveals distinct targeting mechanisms for master transcription factor GATA1
Blood
·
21 Apr 2022
·
doi:10.1182/blood.2021013753
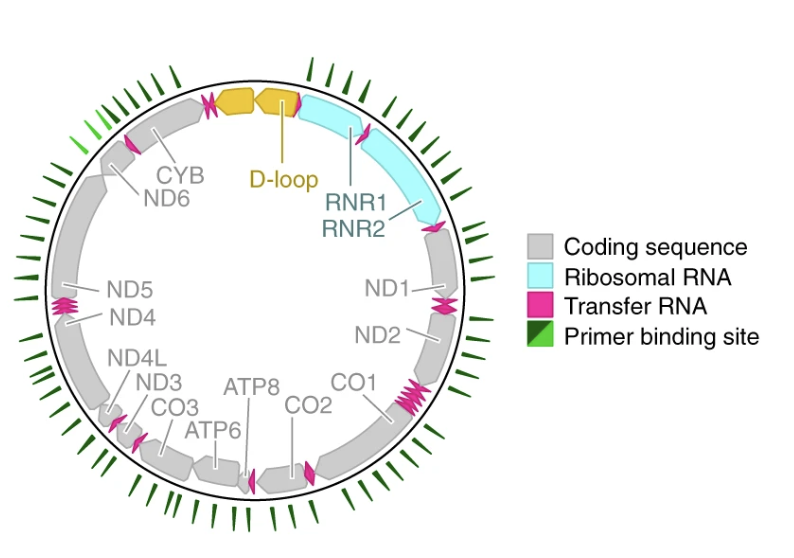
Mitochondrial variant enrichment from high-throughput single-cell RNA sequencing resolves clonal populations
Nature Biotechnology
·
24 Feb 2022
·
doi:10.1038/s41587-022-01210-8
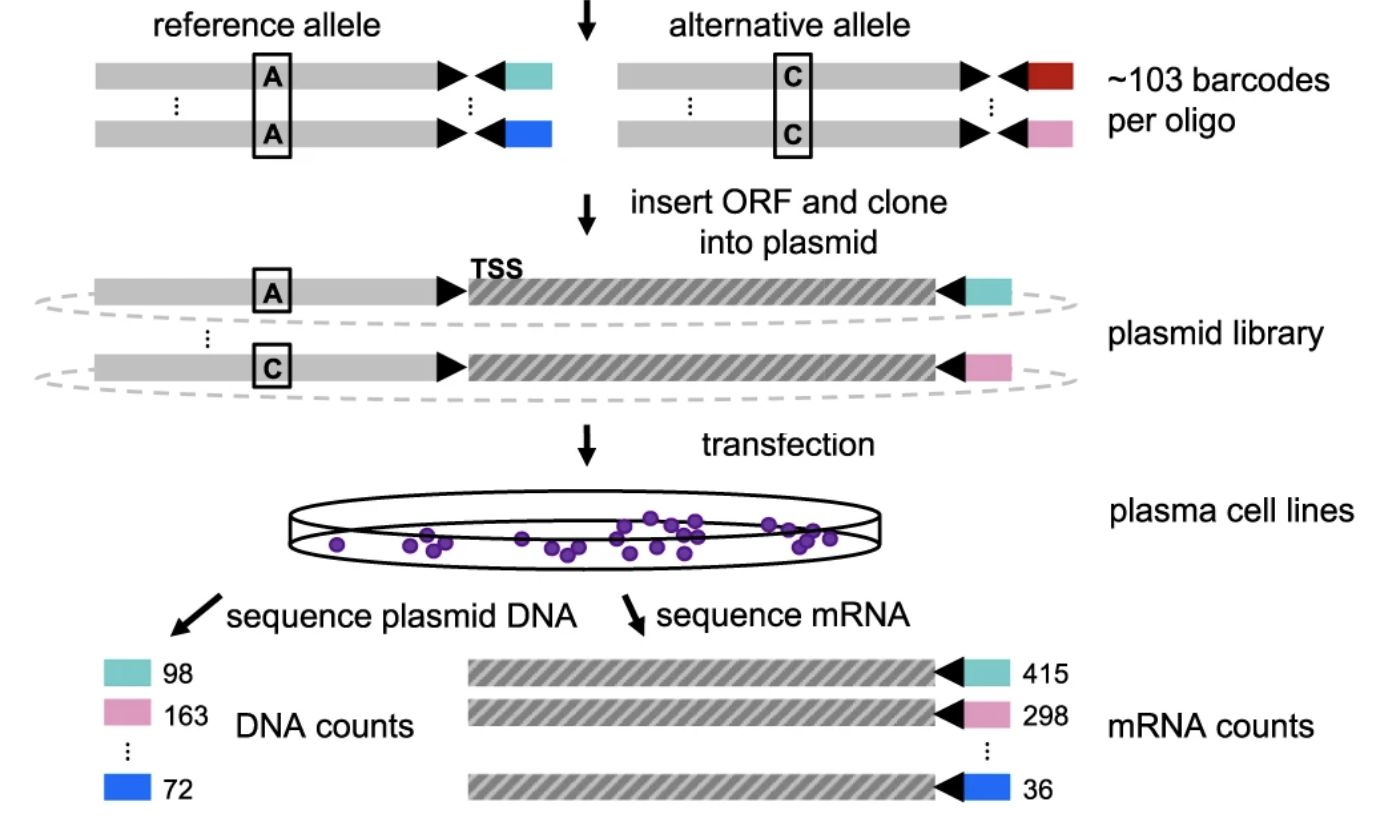
Functional dissection of inherited non-coding variation influencing multiple myeloma risk
Nature Communications
·
10 Jan 2022
·
doi:10.1038/s41467-021-27666-x
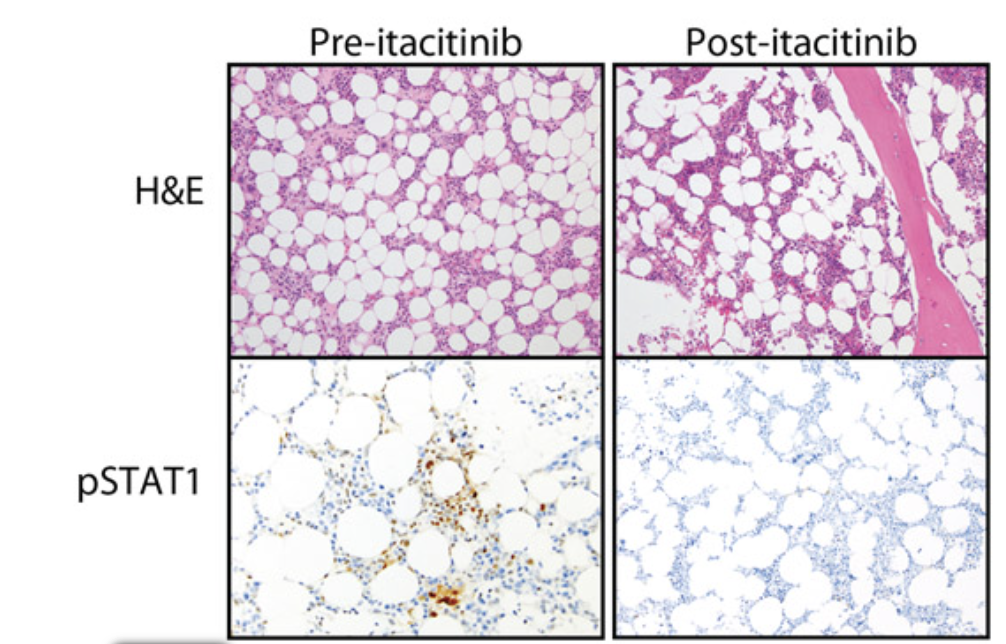
JAK inhibition in a patient with a STAT1 gain-of-function variant reveals STAT1 dysregulation as a common feature of aplastic anemia
Med
·
01 Jan 2022
·
doi:10.1016/j.medj.2021.12.003
2021
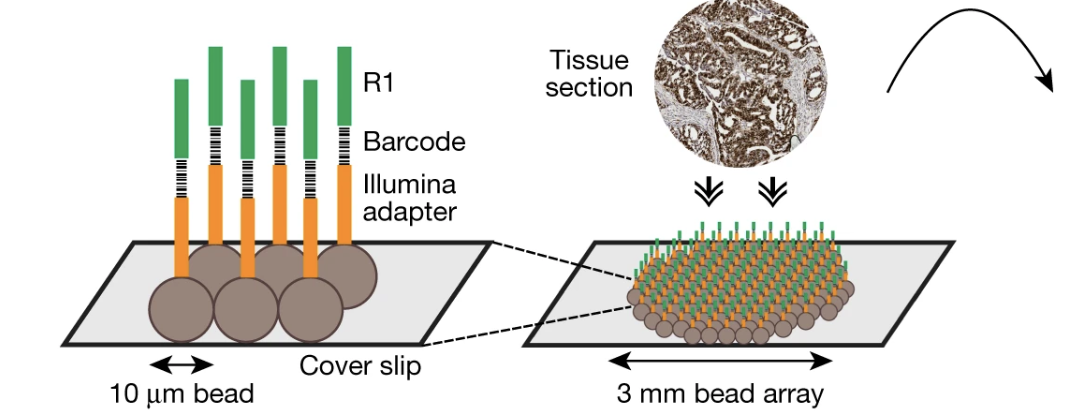
Spatial genomics enables multi-modal study of clonal heterogeneity in tissues
Nature
·
15 Dec 2021
·
doi:10.1038/s41586-021-04217-4
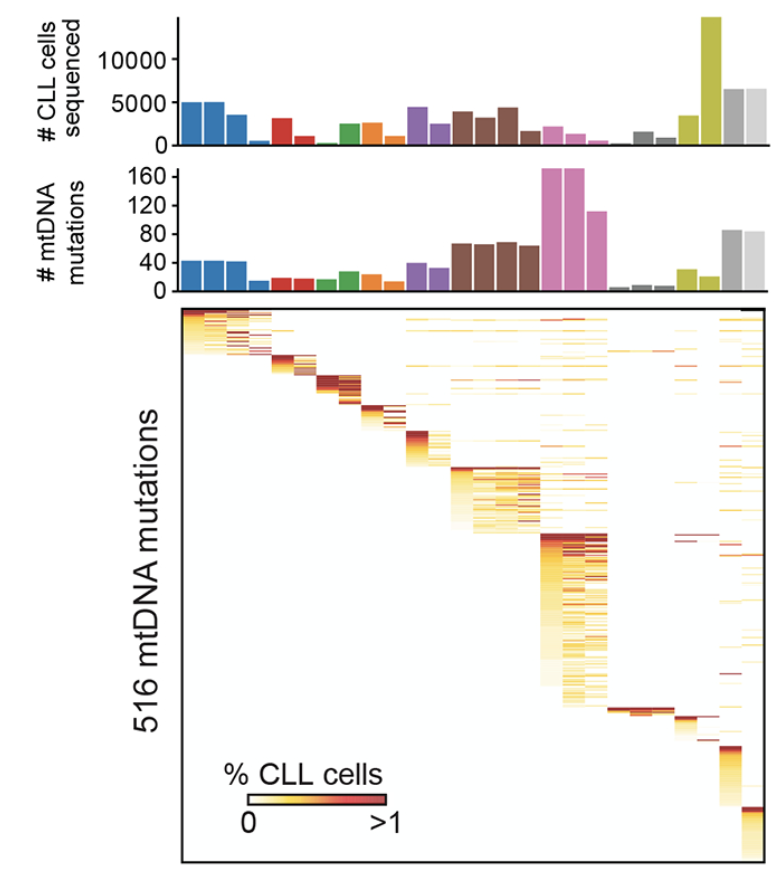
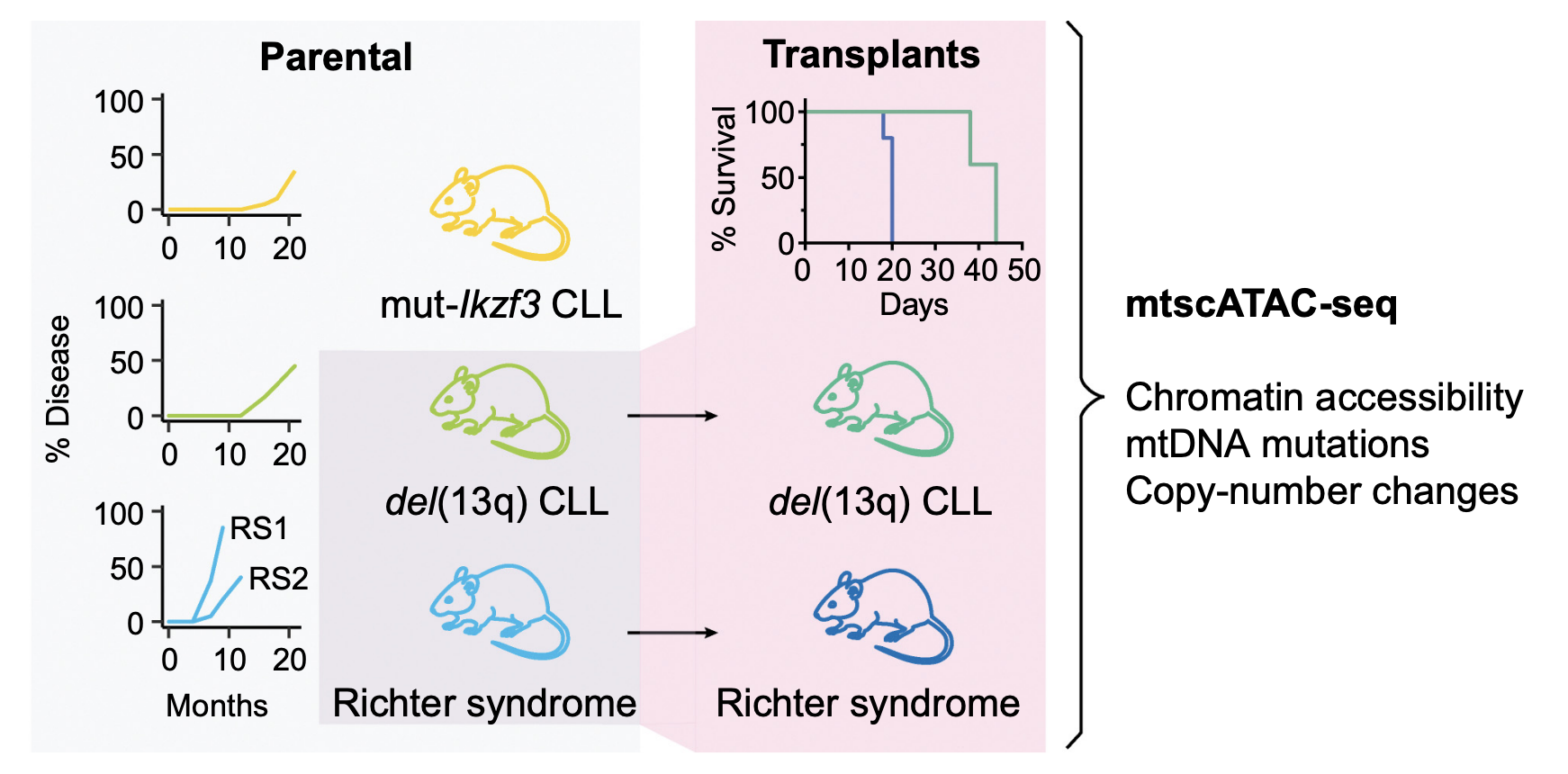
Longitudinal Single-Cell Dynamics of Chromatin Accessibility and Mitochondrial Mutations in Chronic Lymphocytic Leukemia Mirror Disease History
Cancer Discovery
·
01 Dec 2021
·
doi:10.1158/2159-8290.CD-21-0276
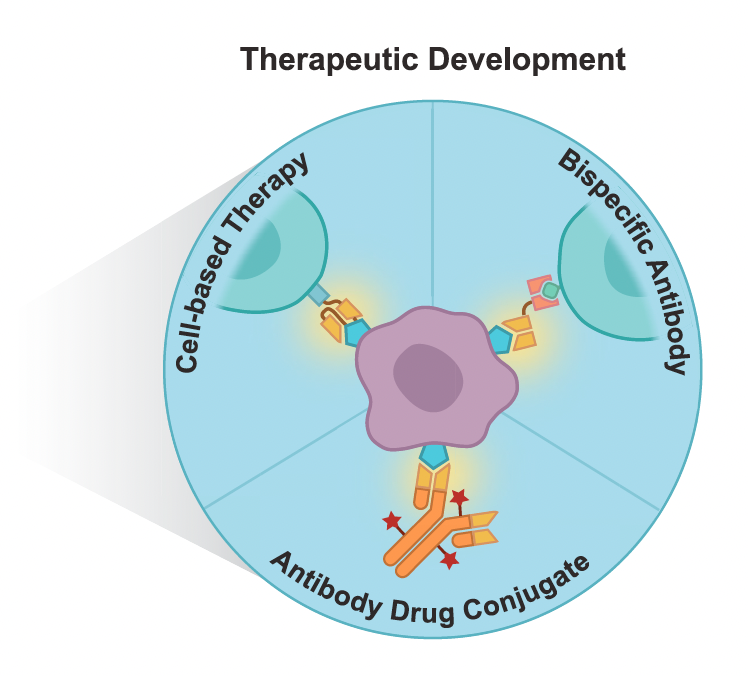
Charting the tumor antigen maps drawn by single-cell genomics
Cancer Cell
·
01 Dec 2021
·
doi:10.1016/j.ccell.2021.11.005
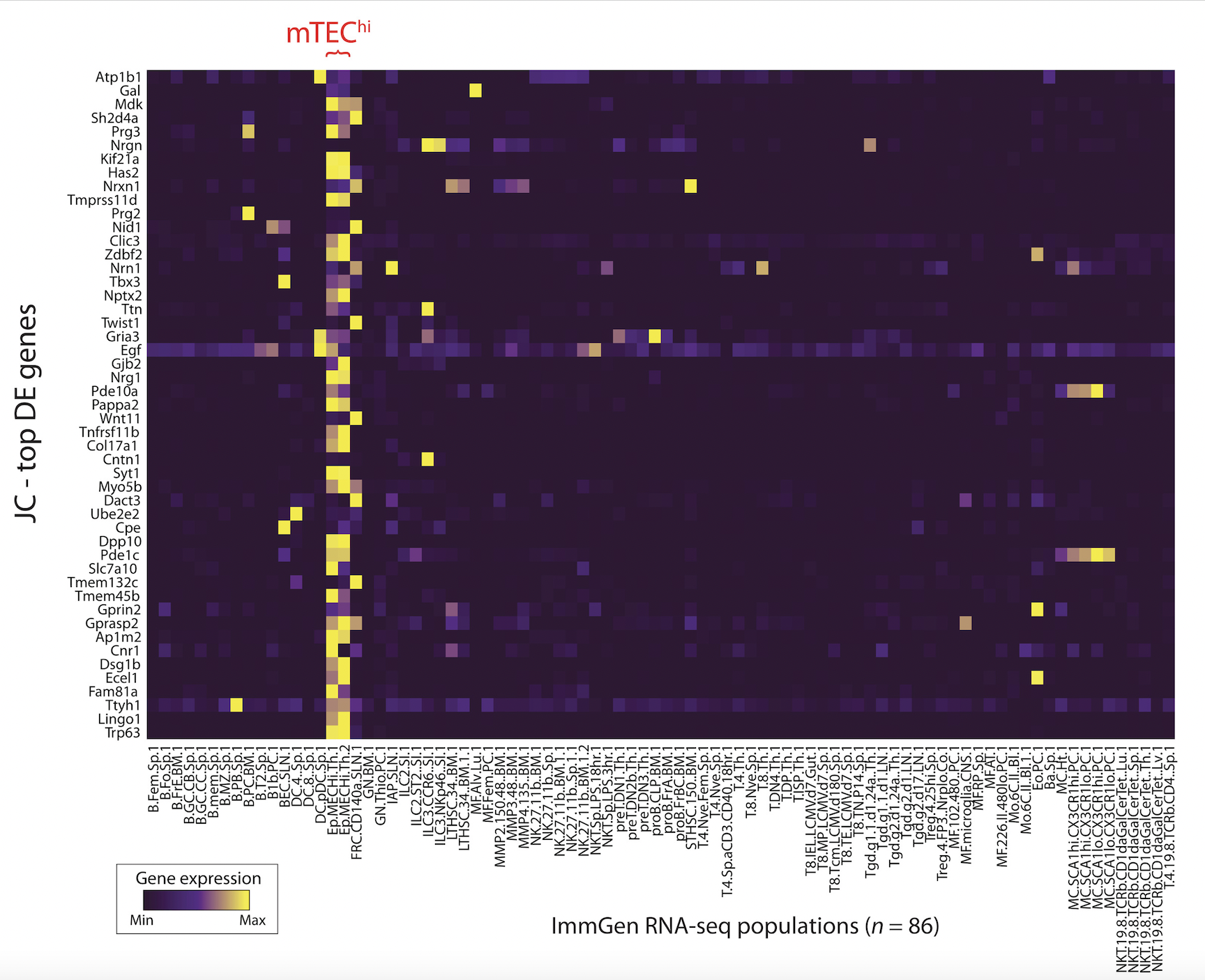
Single-cell multiomics defines tolerogenic extrathymic Aire-expressing populations with unique homology to thymic epithelium
Science Immunology
·
26 Nov 2021
·
doi:10.1126/sciimmunol.abl5053
Single-cell chromatin state analysis with Signac
Nature Methods
·
01 Nov 2021
·
doi:10.1038/s41592-021-01282-5
Integrated single-cell transcriptomics and epigenomics reveals strong germinal center–associated etiology of autoimmune risk loci
Science Immunology
·
29 Oct 2021
·
doi:10.1126/sciimmunol.abh3768
Single-cell profiling of proteins and chromatin accessibility using PHAGE-ATAC
Nature Biotechnology
·
21 Oct 2021
·
doi:10.1038/s41587-021-01065-5
Transient dendritic cell activation diversifies the T cell response to acute infection
Cold Spring Harbor Laboratory
·
27 Sep 2021
·
doi:10.1101/2021.09.26.461821

Scalable, multimodal profiling of chromatin accessibility, gene expression and protein levels in single cells
Nature Biotechnology
·
03 Jun 2021
·
doi:10.1038/s41587-021-00927-2
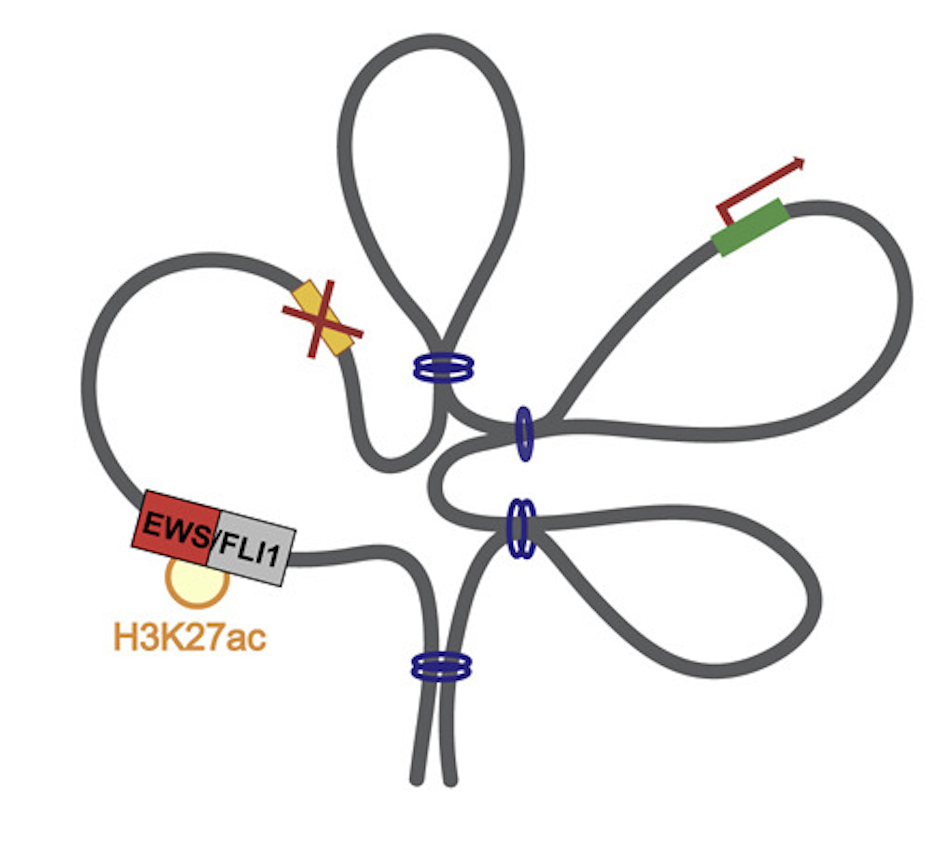
STAG2 loss rewires oncogenic and developmental programs to promote metastasis in Ewing sarcoma
Cancer Cell
·
01 Jun 2021
·
doi:10.1016/j.ccell.2021.05.007
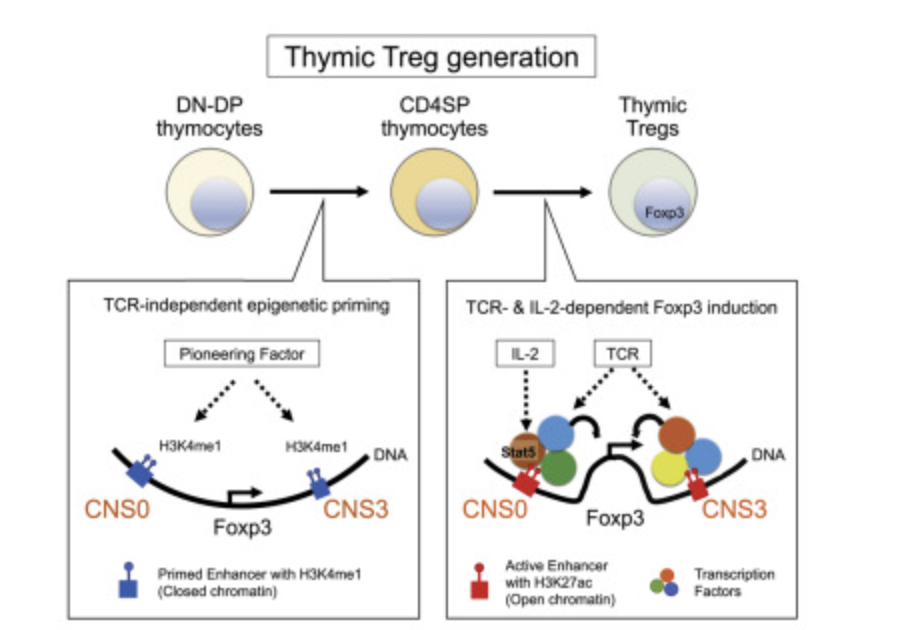
Distinct Foxp3 enhancer elements coordinate development, maintenance, and function of regulatory T cells
Immunity
·
01 May 2021
·
doi:10.1016/j.immuni.2021.04.005
2020
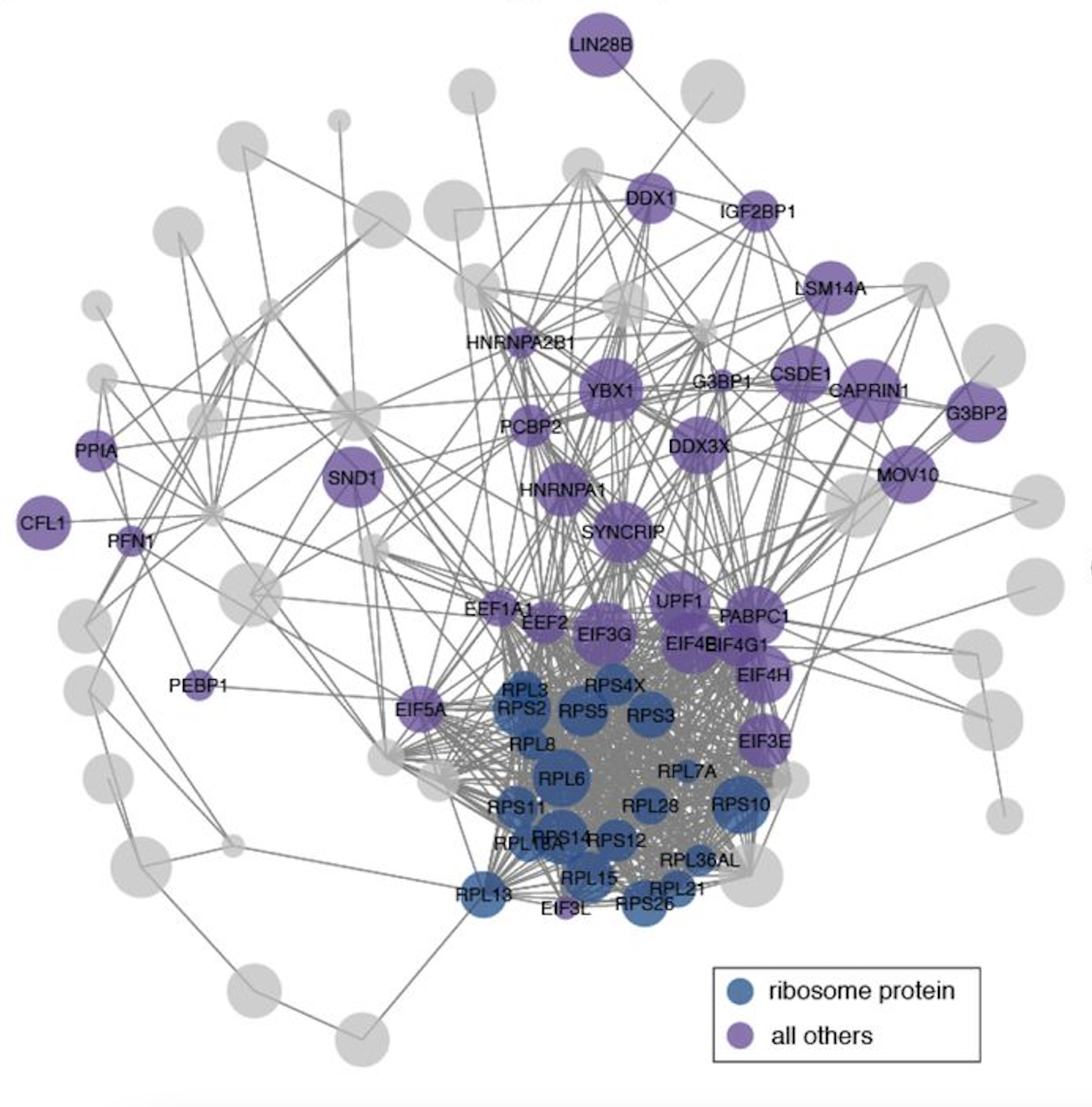
The SARS-CoV-2 RNA–protein interactome in infected human cells
Nature Microbiology
·
21 Dec 2020
·
doi:10.1038/s41564-020-00846-z
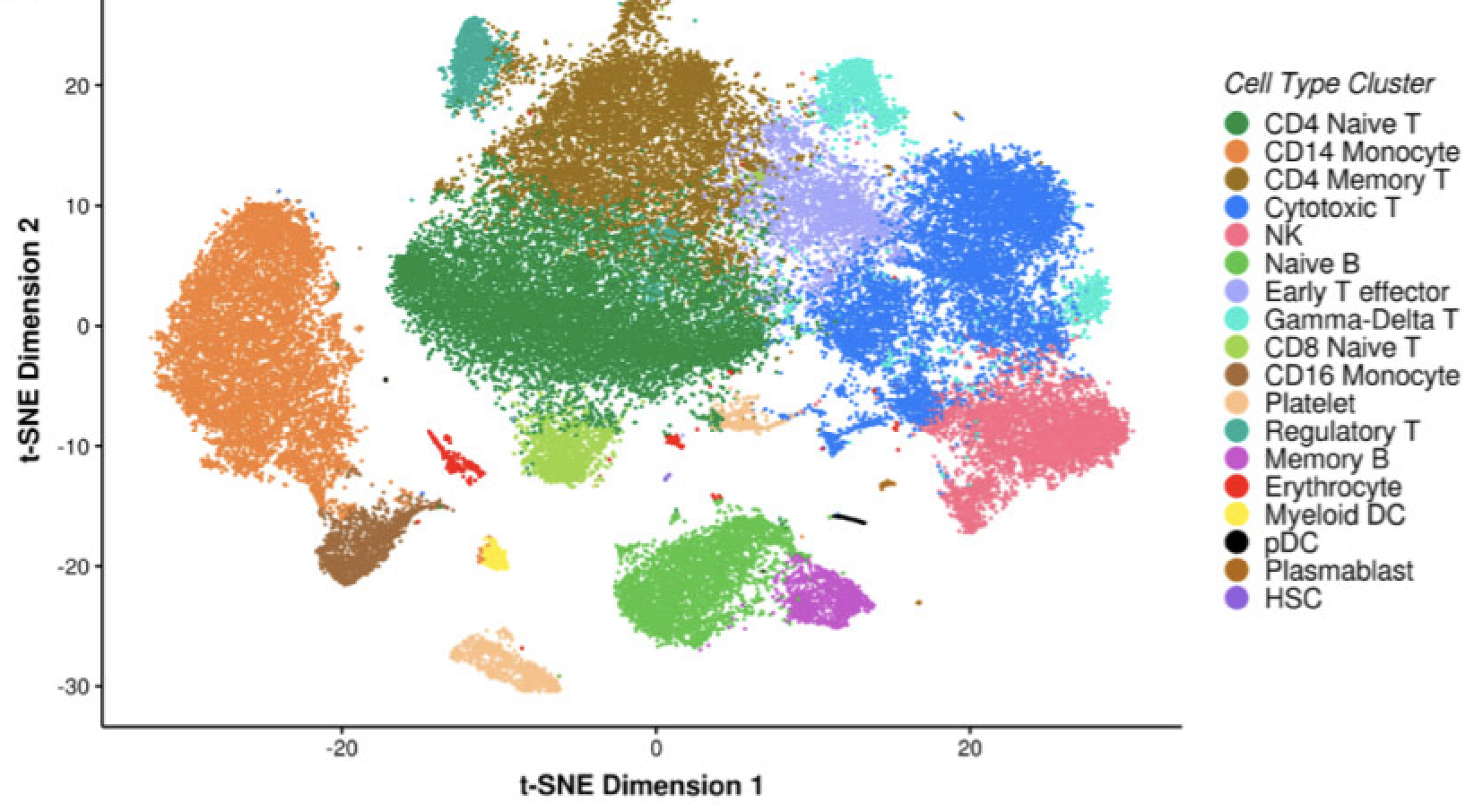
Single Cell Transcriptomics Implicate Novel Monocyte and T Cell Immune Dysregulation in Sarcoidosis
Frontiers in Immunology
·
08 Dec 2020
·
doi:10.3389/fimmu.2020.567342
Chromatin Potential Identified by Shared Single-Cell Profiling of RNA and Chromatin
Cell
·
01 Nov 2020
·
doi:10.1016/j.cell.2020.09.056
Purifying Selection against Pathogenic Mitochondrial DNA in Human T Cells
New England Journal of Medicine
·
15 Oct 2020
·
doi:10.1056/NEJMoa2001265
Inherited myeloproliferative neoplasm risk affects haematopoietic stem cells
Nature
·
14 Oct 2020
·
doi:10.1038/s41586-020-2786-7
An old BATF’s new T-ricks
Nature Immunology
·
28 Sep 2020
·
doi:10.1038/s41590-020-0796-0
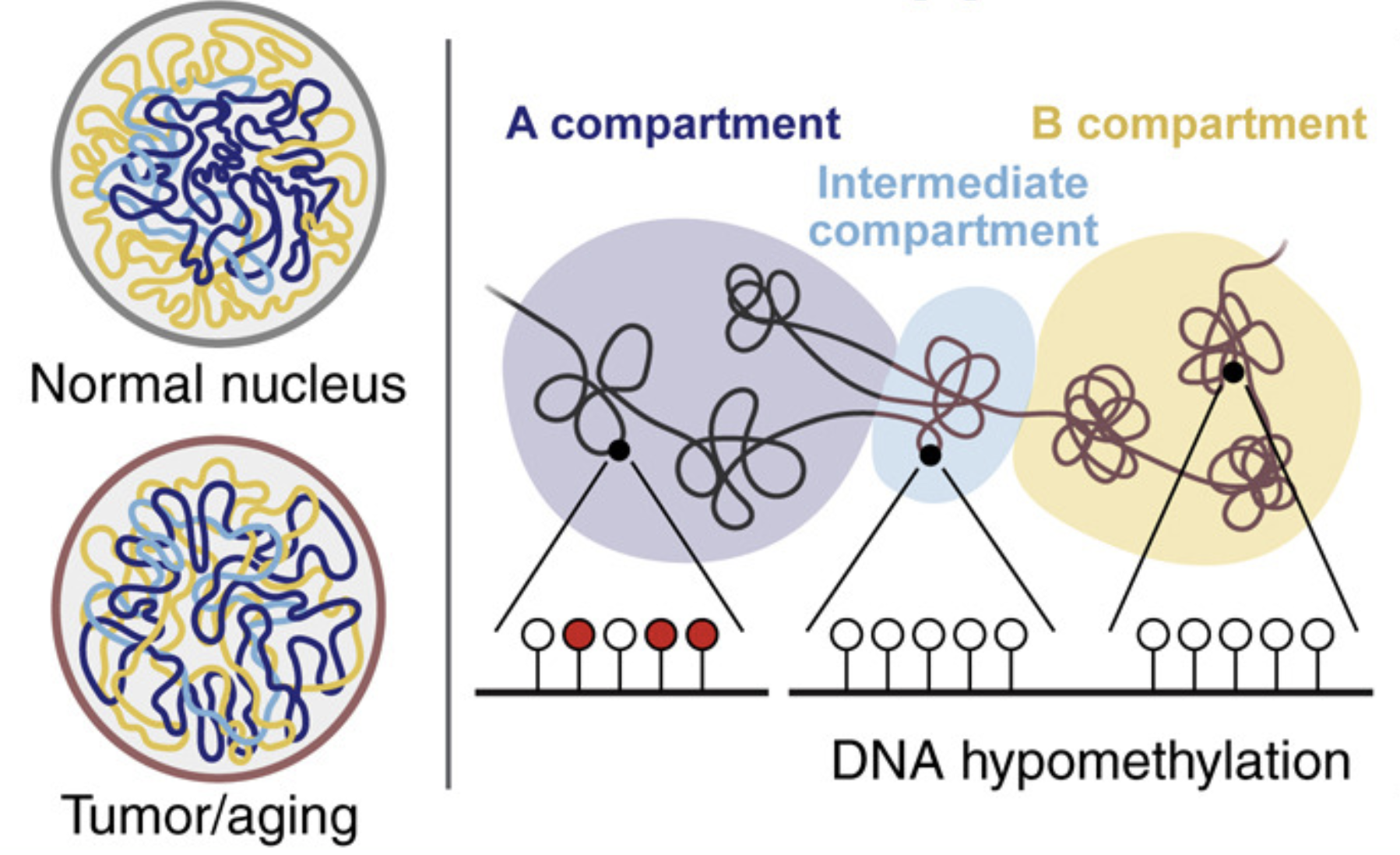
Large-Scale Topological Changes Restrain Malignant Progression in Colorectal Cancer
Cell
·
01 Sep 2020
·
doi:10.1016/j.cell.2020.07.030
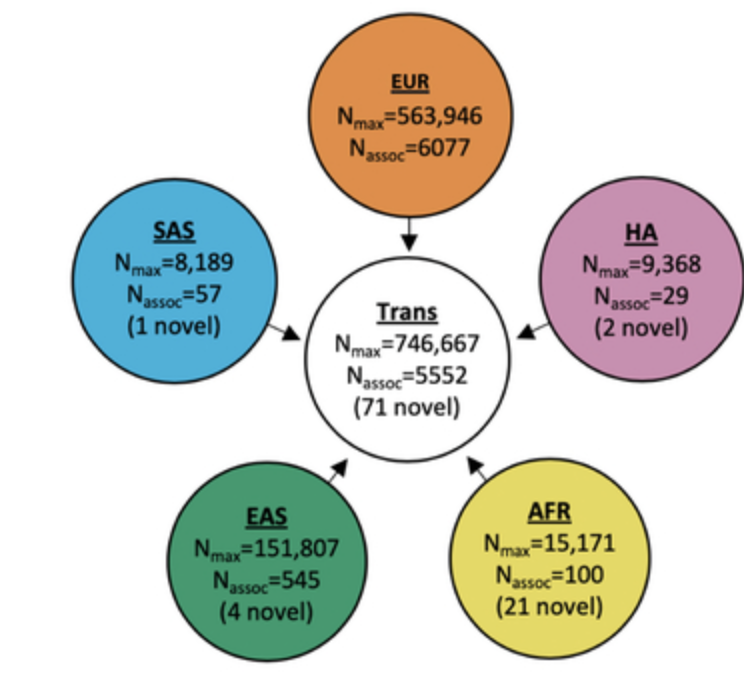
Trans-ethnic and Ancestry-Specific Blood-Cell Genetics in 746,667 Individuals from 5 Global Populations
Cell
·
01 Sep 2020
·
doi:10.1016/j.cell.2020.06.045
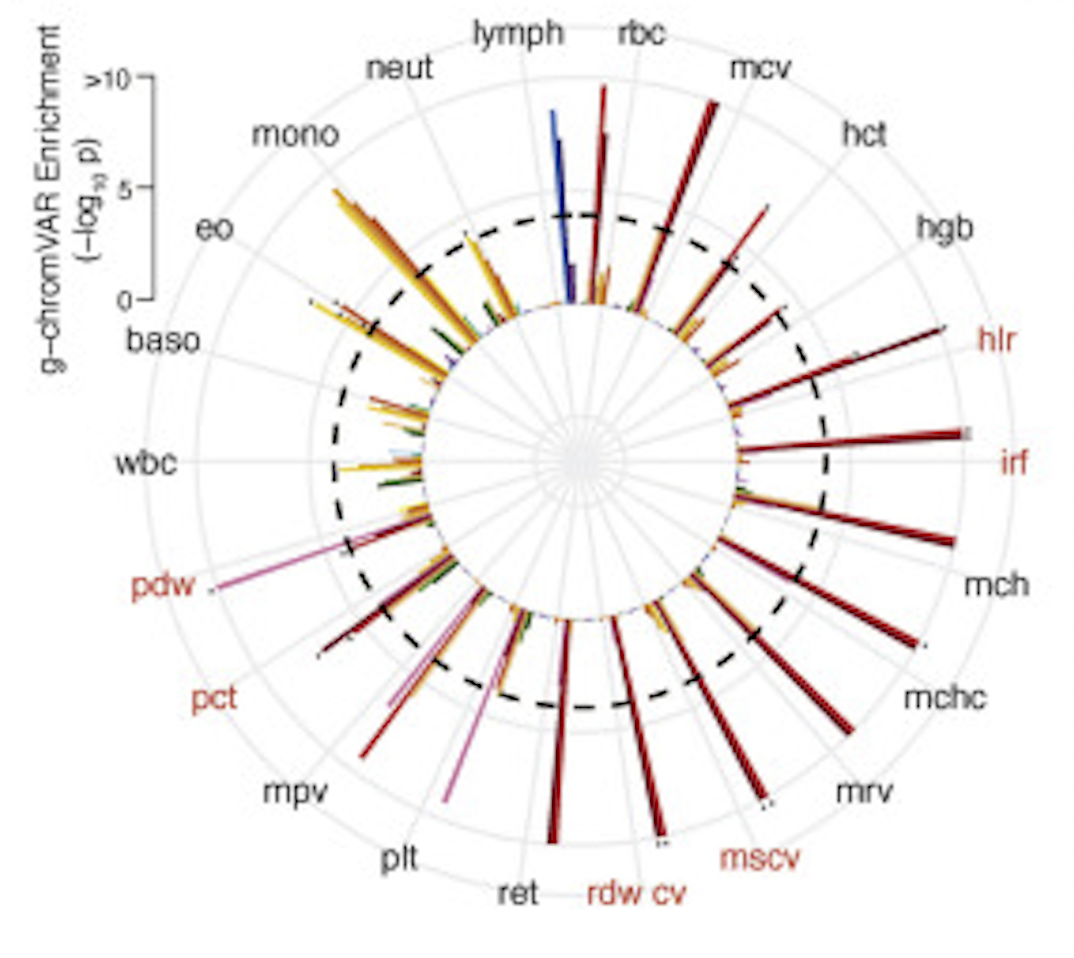
The Polygenic and Monogenic Basis of Blood Traits and Diseases
Cell
·
01 Sep 2020
·
doi:10.1016/j.cell.2020.08.008
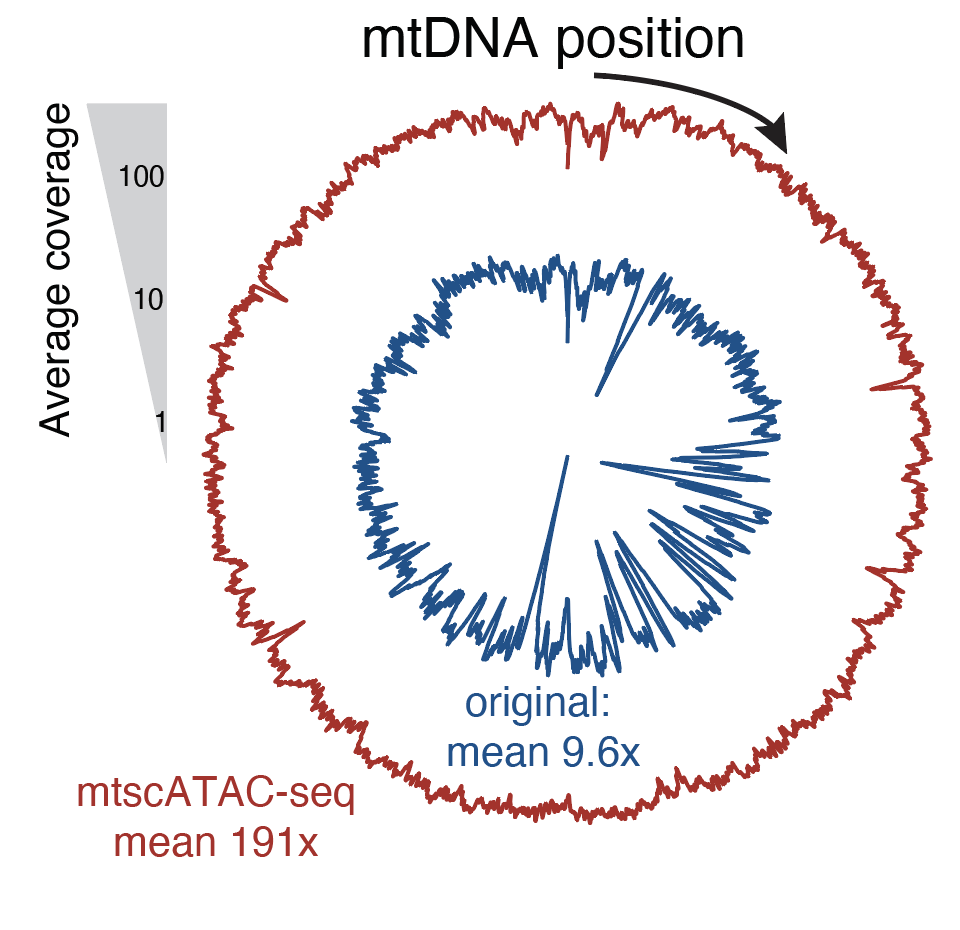
Massively parallel single-cell mitochondrial DNA genotyping and chromatin profiling
Nature Biotechnology
·
12 Aug 2020
·
doi:10.1038/s41587-020-0645-6
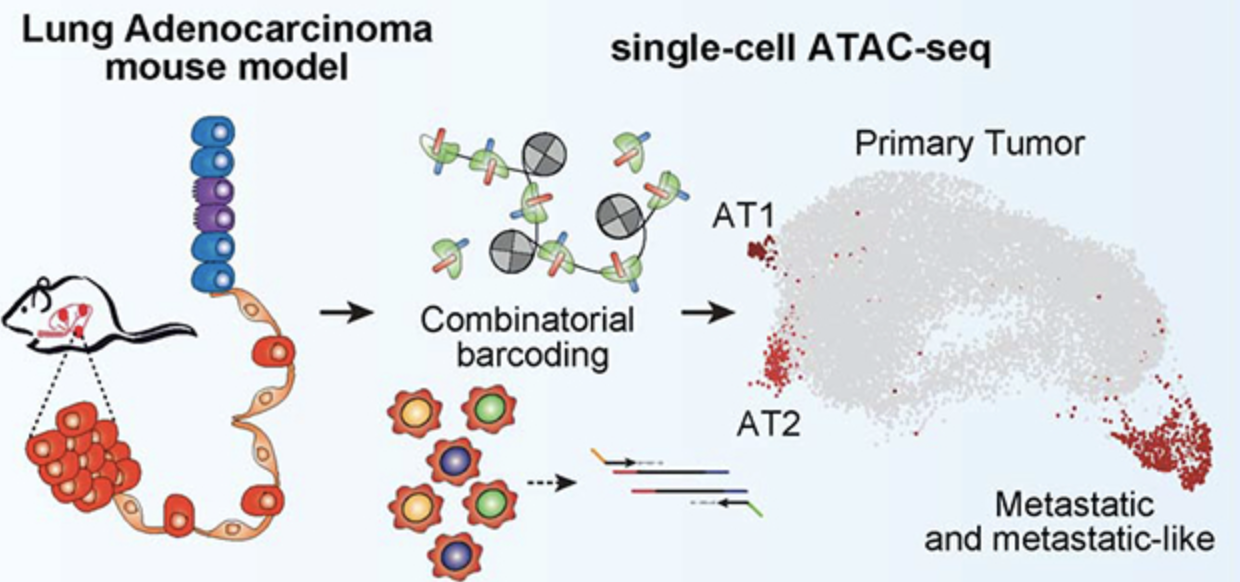
Epigenomic State Transitions Characterize Tumor Progression in Mouse Lung Adenocarcinoma
Cancer Cell
·
01 Aug 2020
·
doi:10.1016/j.ccell.2020.06.006

ImmGen at 15
Nature Immunology
·
23 Jun 2020
·
doi:10.1038/s41590-020-0687-4
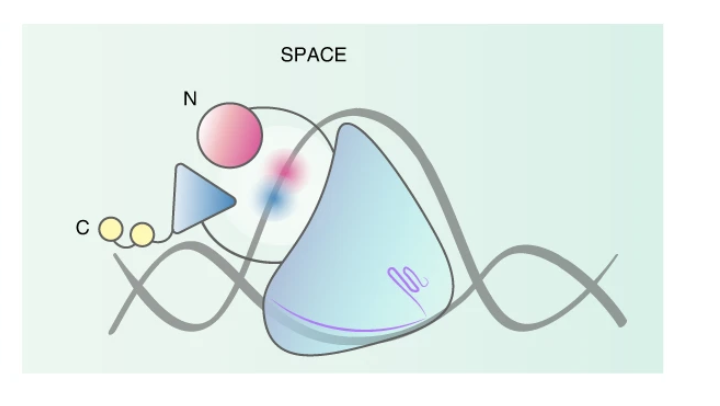
A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing
Nature Biotechnology
·
01 Jun 2020
·
doi:10.1038/s41587-020-0535-y
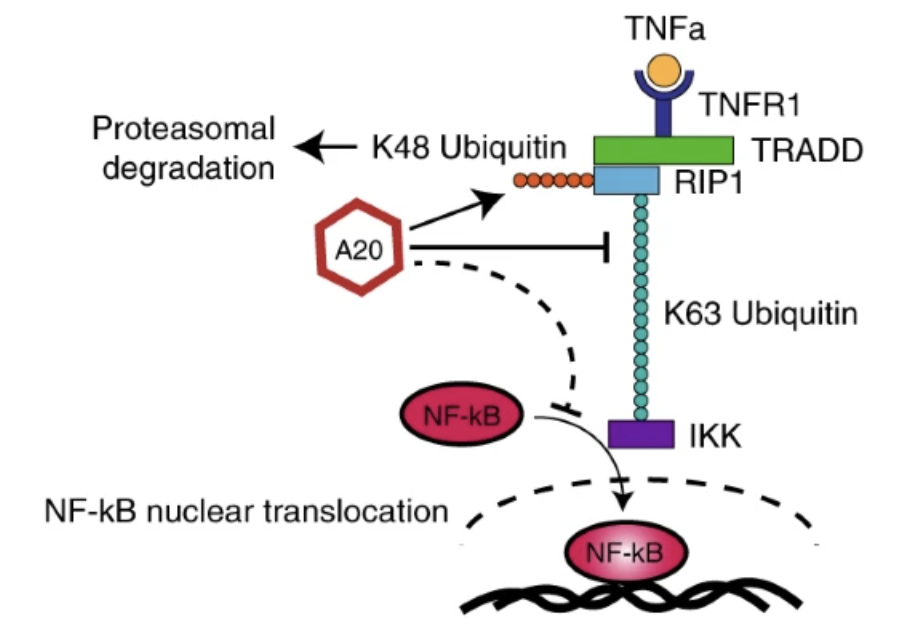
Prioritizing disease and trait causal variants at the TNFAIP3 locus using functional and genomic features
Nature Communications
·
06 Mar 2020
·
doi:10.1038/s41467-020-15022-4
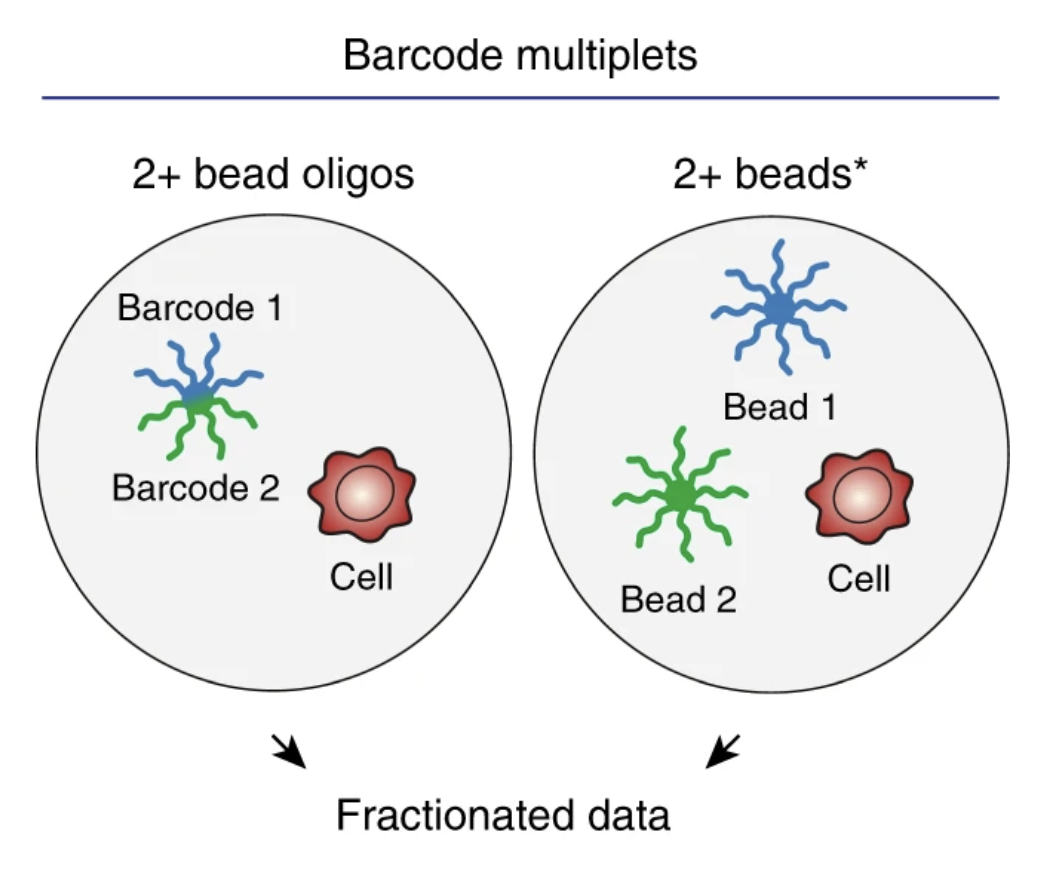
Inference and effects of barcode multiplets in droplet-based single-cell assays
Nature Communications
·
13 Feb 2020
·
doi:10.1038/s41467-020-14667-5
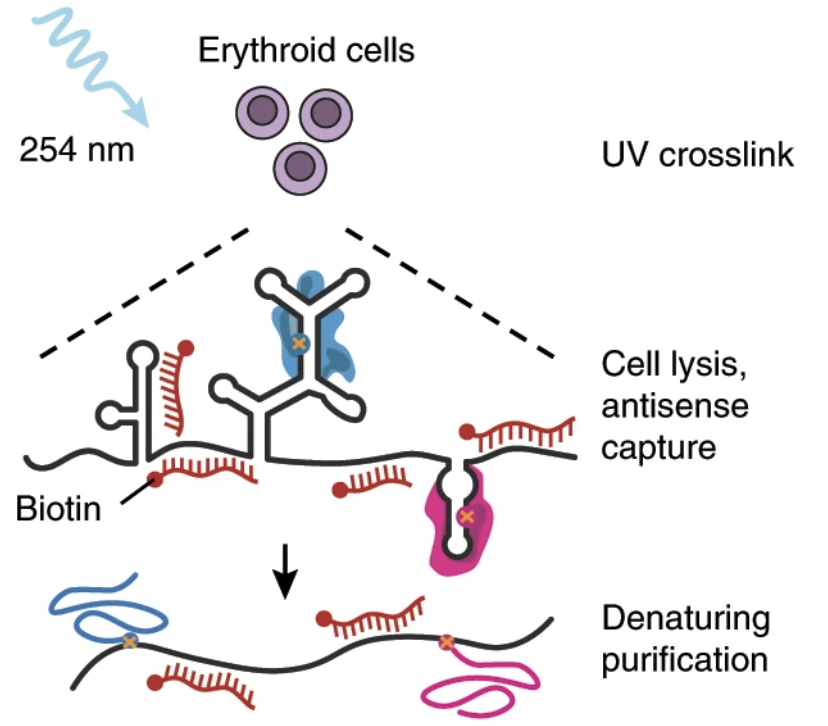
Control of human hemoglobin switching by LIN28B-mediated regulation of BCL11A translation
Nature Genetics
·
20 Jan 2020
·
doi:10.1038/s41588-019-0568-7
2019
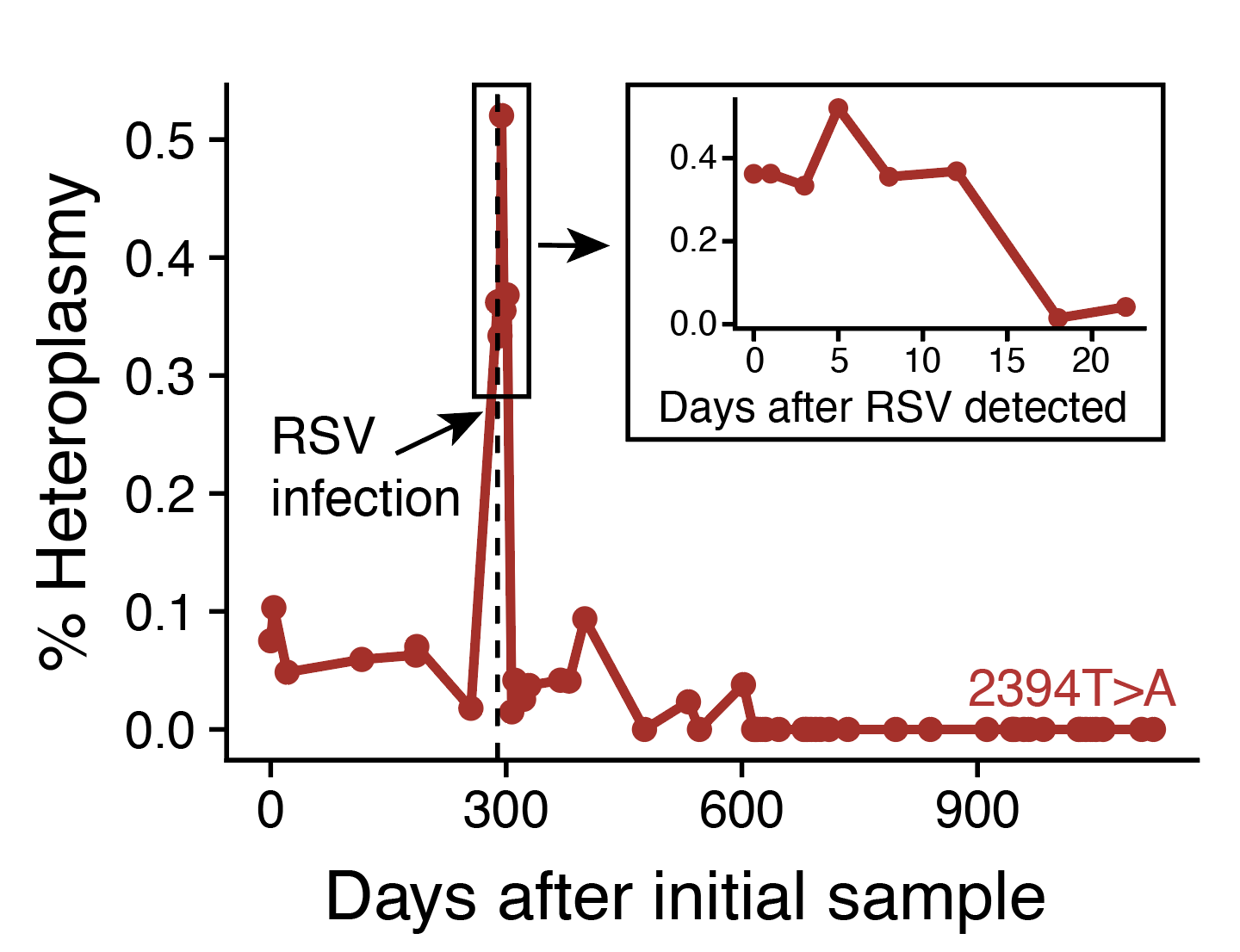
Longitudinal assessment of clonal mosaicism in human hematopoiesis via mitochondrial mutation tracking
Blood Advances
·
16 Dec 2019
·
doi:10.1182/bloodadvances.2019001196
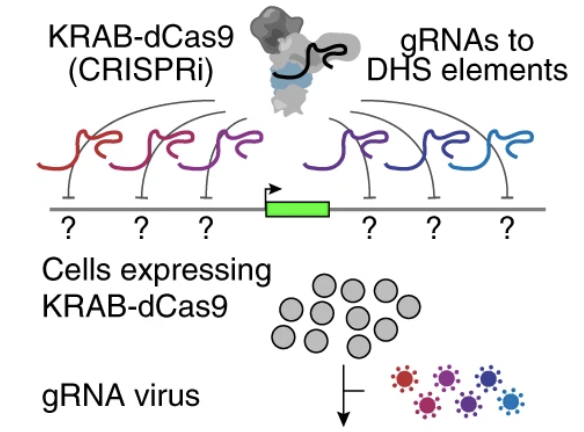
Activity-by-contact model of enhancer–promoter regulation from thousands of CRISPR perturbations
Nature Genetics
·
29 Nov 2019
·
doi:10.1038/s41588-019-0538-0
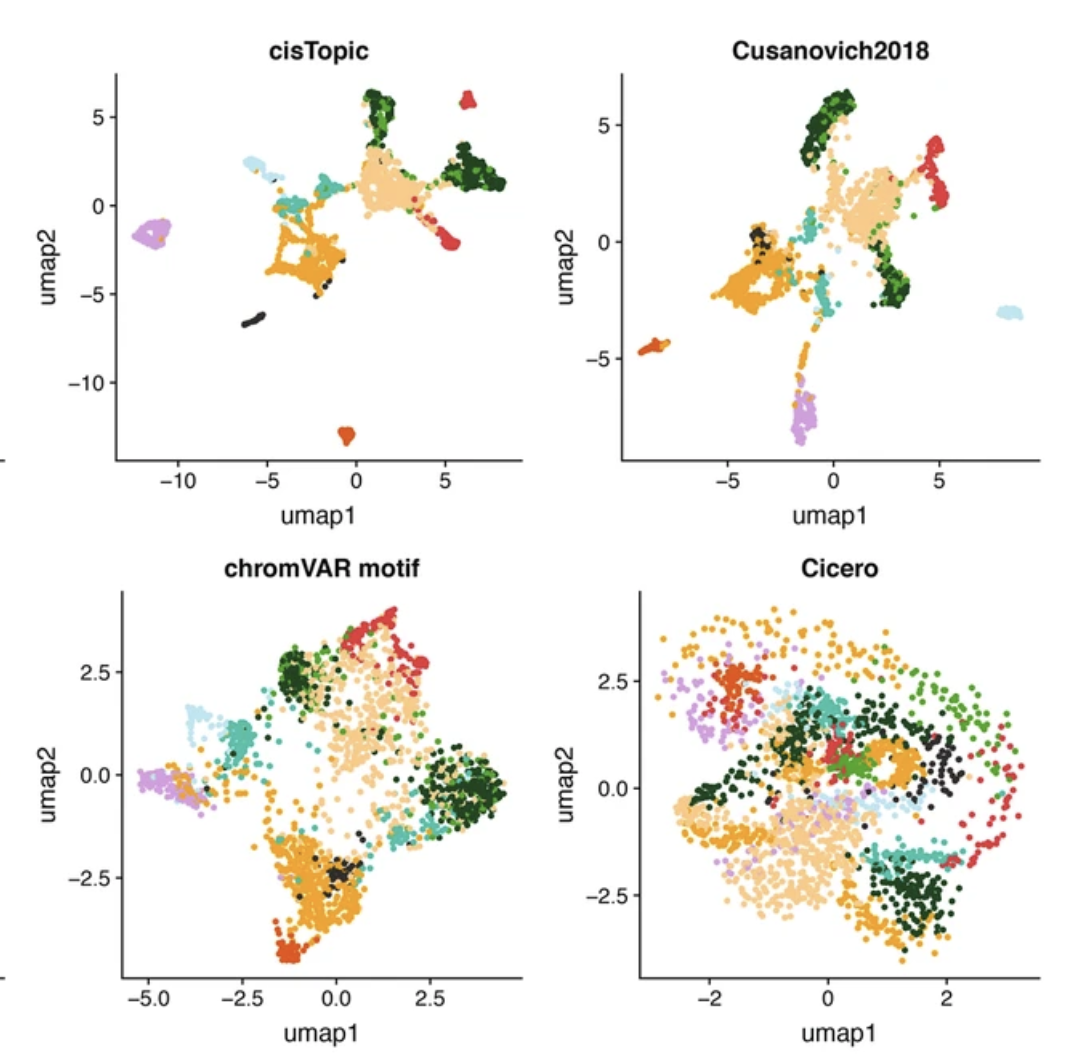
Assessment of computational methods for the analysis of single-cell ATAC-seq data
Genome Biology
·
18 Nov 2019
·
doi:10.1186/s13059-019-1854-5
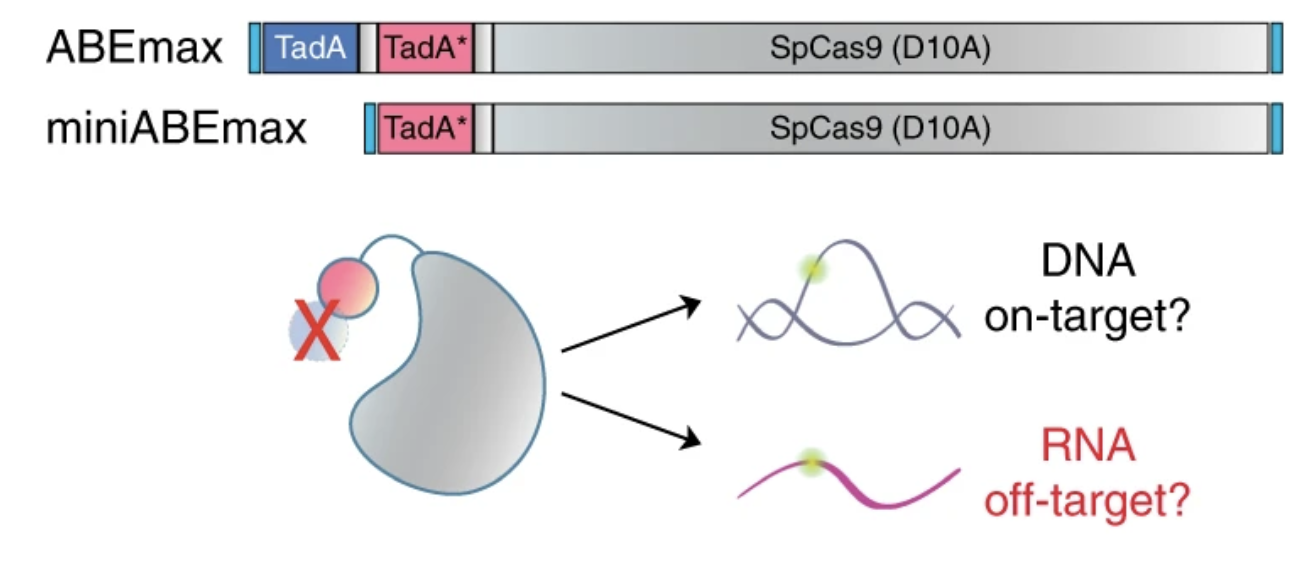
CRISPR DNA base editors with reduced RNA off-target and self-editing activities
Nature Biotechnology
·
01 Sep 2019
·
doi:10.1038/s41587-019-0236-6
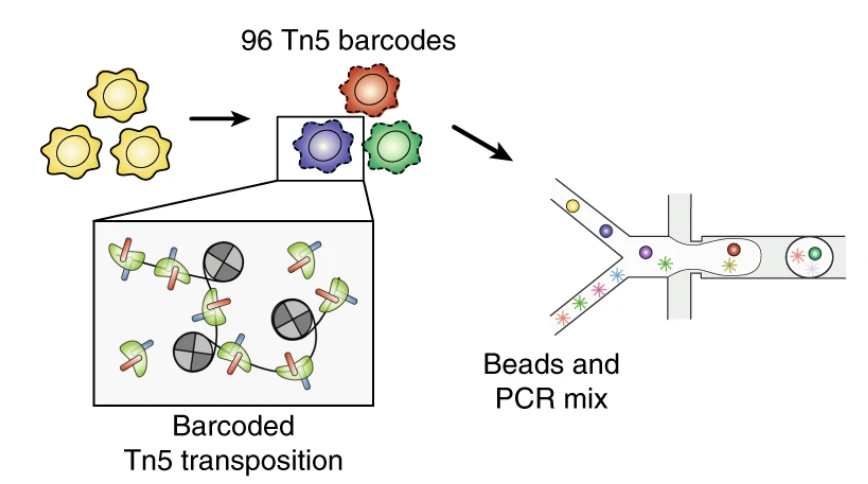
Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility
Nature Biotechnology
·
24 Jun 2019
·
doi:10.1038/s41587-019-0147-6
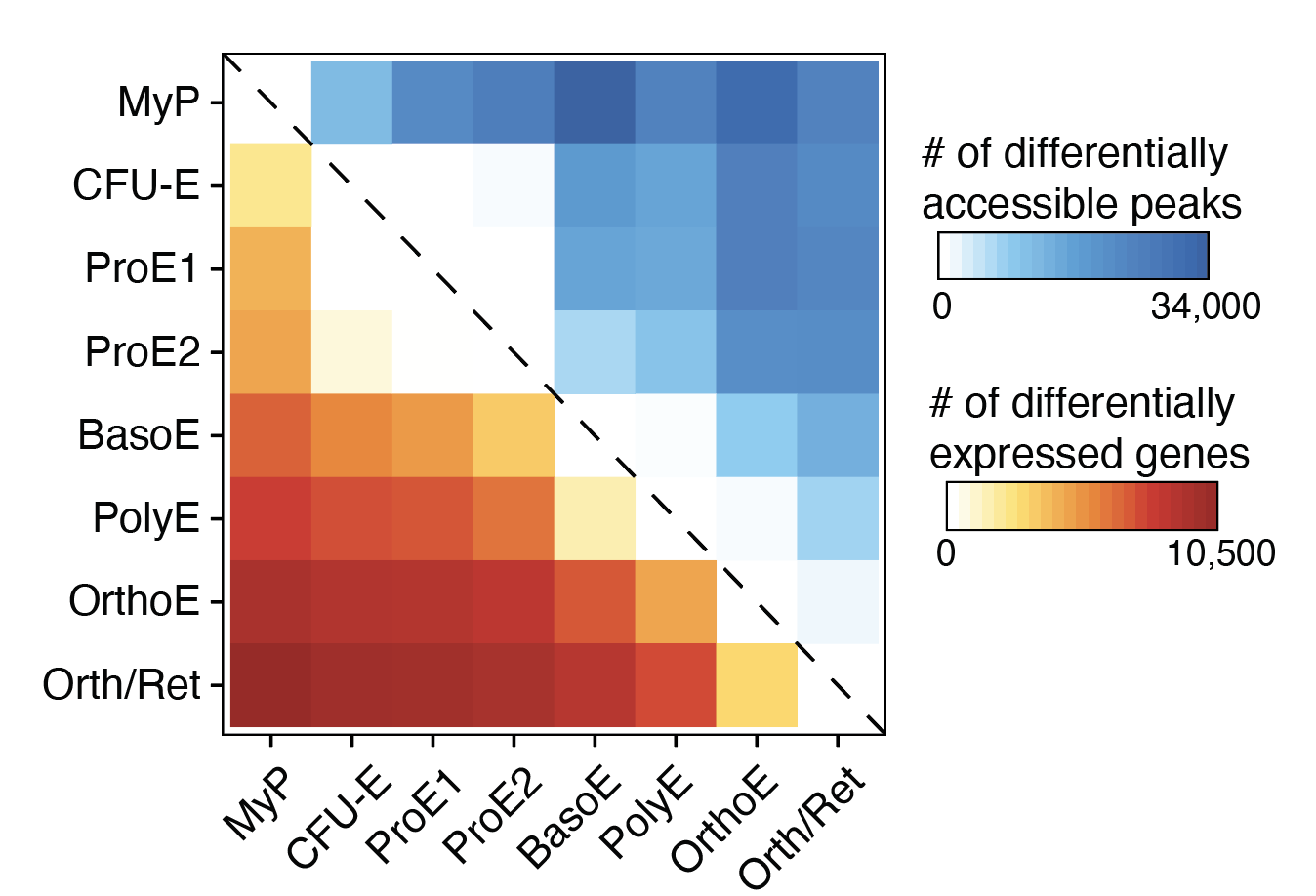
Transcriptional States and Chromatin Accessibility Underlying Human Erythropoiesis
Cell Reports
·
01 Jun 2019
·
doi:10.1016/j.celrep.2019.05.046
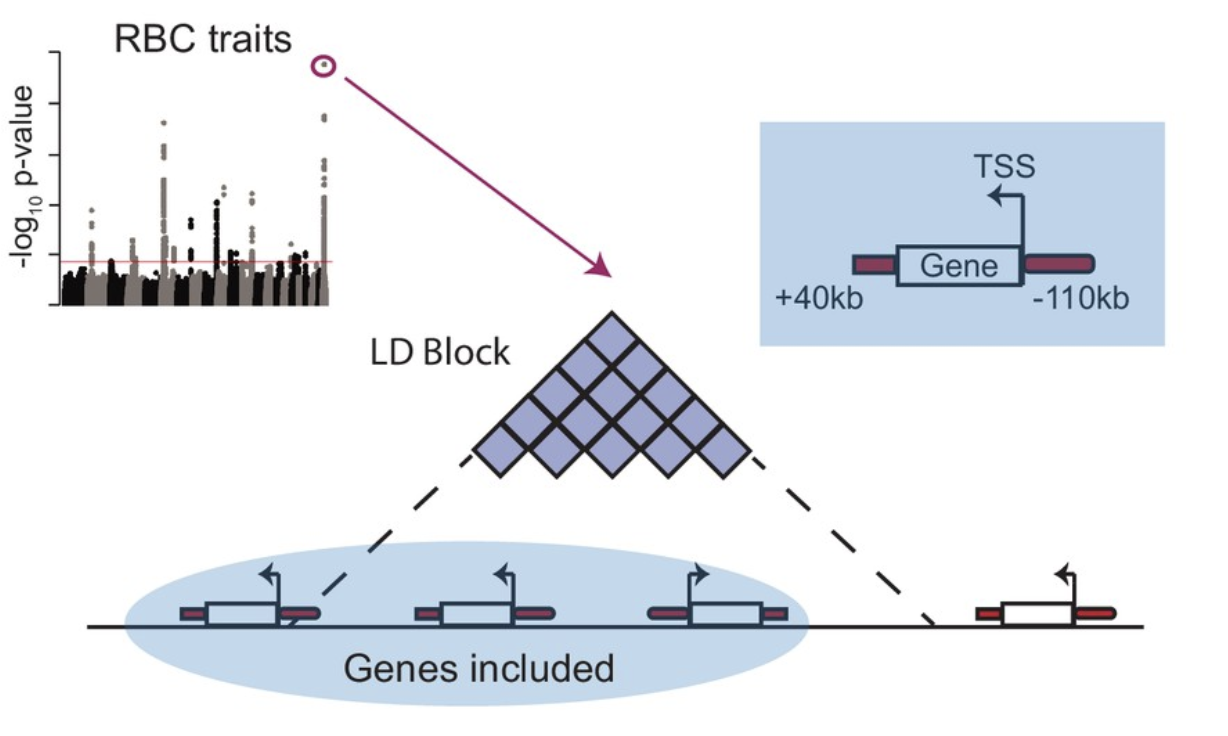
Gene-centric functional dissection of human genetic variation uncovers regulators of hematopoiesis
eLife
·
09 May 2019
·
doi:10.7554/eLife.44080
Single-cell trajectories reconstruction, exploration and mapping of omics data with STREAM
Nature Communications
·
23 Apr 2019
·
doi:10.1038/s41467-019-09670-4
Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors
Nature
·
17 Apr 2019
·
doi:10.1038/s41586-019-1161-z
Impaired human hematopoiesis due to a cryptic intronic GATA1 splicing mutation
Journal of Experimental Medicine
·
26 Mar 2019
·
doi:10.1084/jem.20181625
The ATPase module of mammalian SWI/SNF family complexes mediates subcomplex identity and catalytic activity–independent genomic targeting
Nature Genetics
·
11 Mar 2019
·
doi:10.1038/s41588-019-0363-5
Interrogation of human hematopoiesis at single-cell and single-variant resolution
Nature Genetics
·
11 Mar 2019
·
doi:10.1038/s41588-019-0362-6
Lineage Tracing in Humans Enabled by Mitochondrial Mutations and Single-Cell Genomics
Cell
·
01 Mar 2019
·
doi:10.1016/j.cell.2019.01.022
Heritability of fetal hemoglobin, white cell count, and other clinical traits from a sickle cell disease family cohort
American Journal of Hematology
·
06 Feb 2019
·
doi:10.1002/ajh.25421
The cis-Regulatory Atlas of the Mouse Immune System
Cell
·
01 Feb 2019
·
doi:10.1016/j.cell.2018.12.036
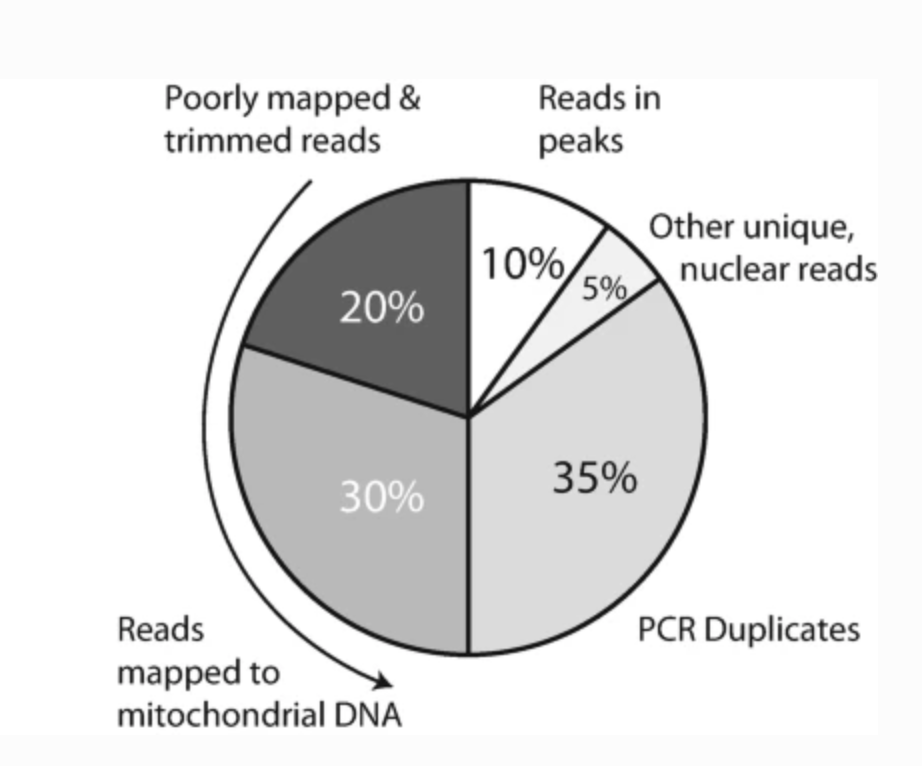
Preprocessing and Computational Analysis of Single-Cell Epigenomic Datasets
Methods in Molecular Biology
·
01 Jan 2019
·
doi:10.1007/978-1-4939-9057-3_13
2018
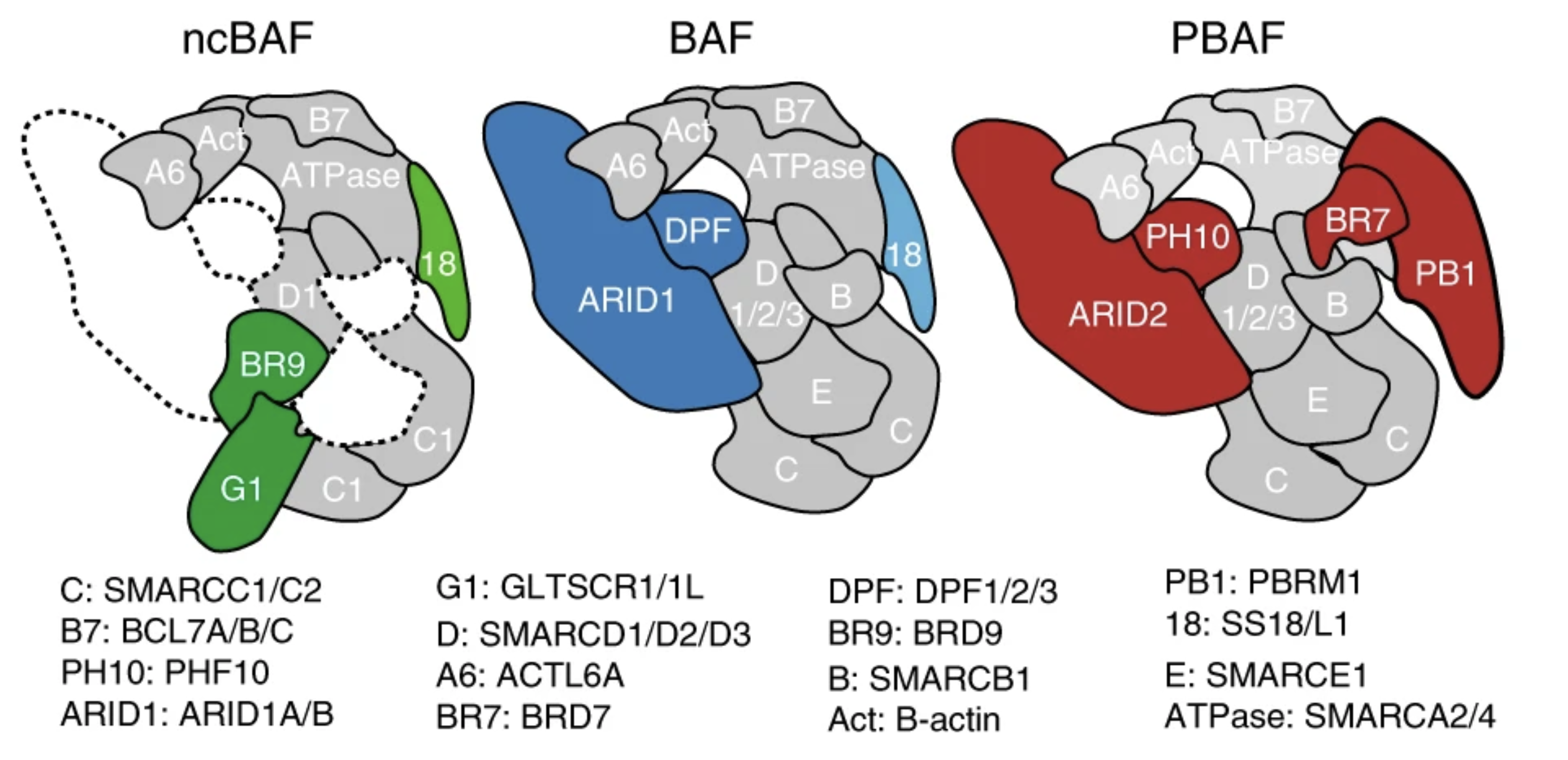
A non-canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation
Nature Cell Biology
·
05 Nov 2018
·
doi:10.1038/s41556-018-0221-1
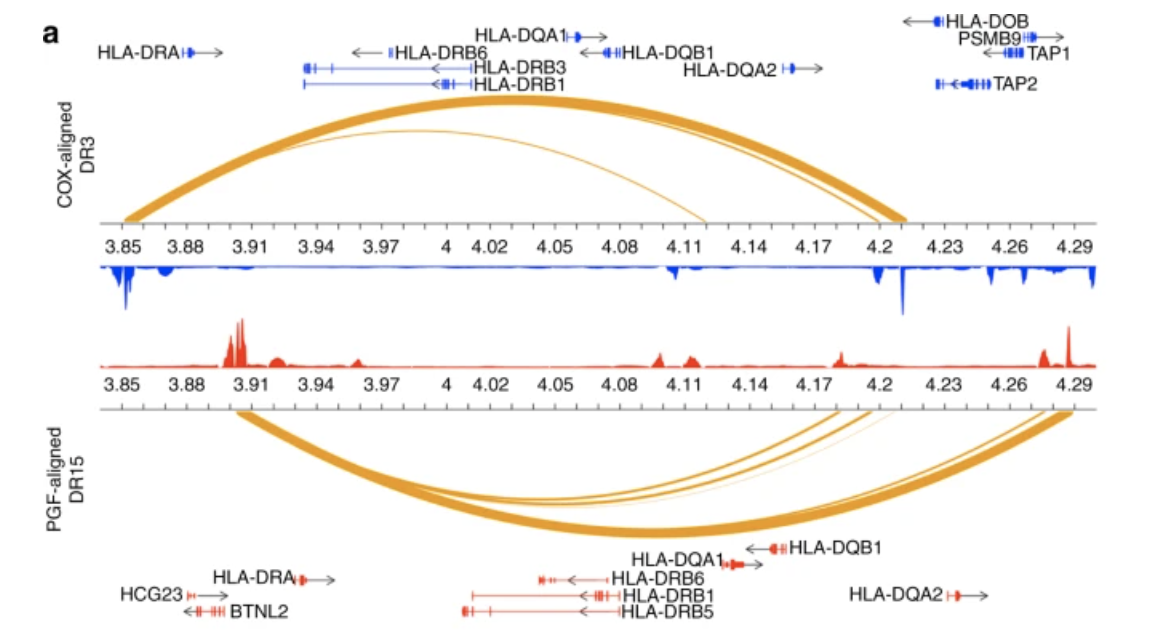
Enhancer histone-QTLs are enriched on autoimmune risk haplotypes and influence gene expression within chromatin networks
Nature Communications
·
25 Jul 2018
·
doi:10.1038/s41467-018-05328-9
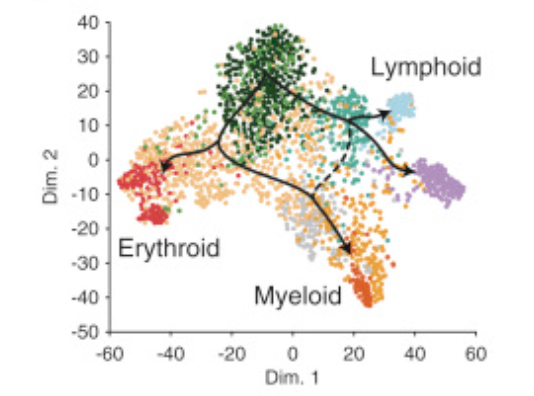
Integrated Single-Cell Analysis Maps the Continuous Regulatory Landscape of Human Hematopoietic Differentiation
Cell
·
01 May 2018
·
doi:10.1016/j.cell.2018.03.074
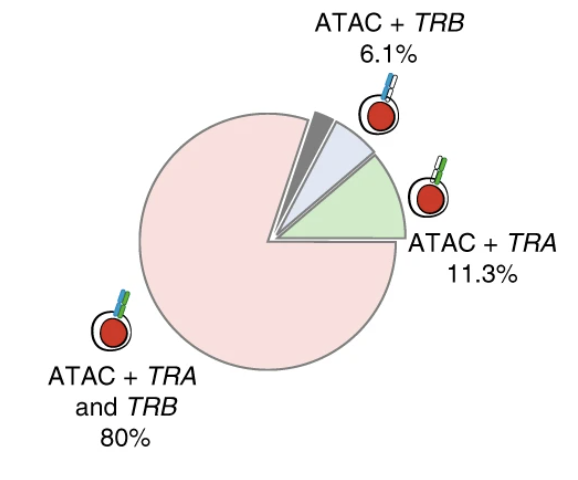
Transcript-indexed ATAC-seq for precision immune profiling
Nature Medicine
·
23 Apr 2018
·
doi:10.1038/S41591-018-0008-8
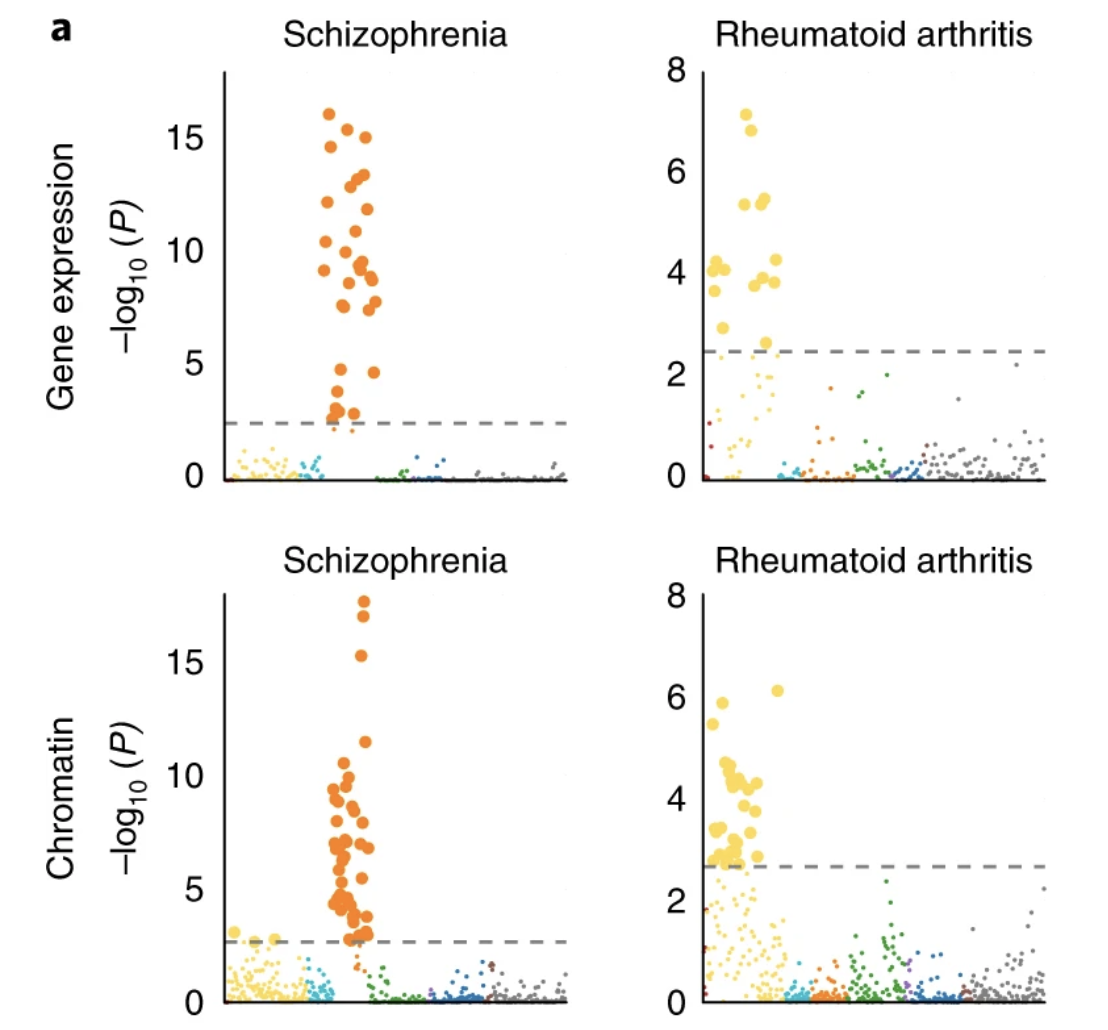
Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types
Nature Genetics
·
01 Apr 2018
·
doi:10.1038/s41588-018-0081-4
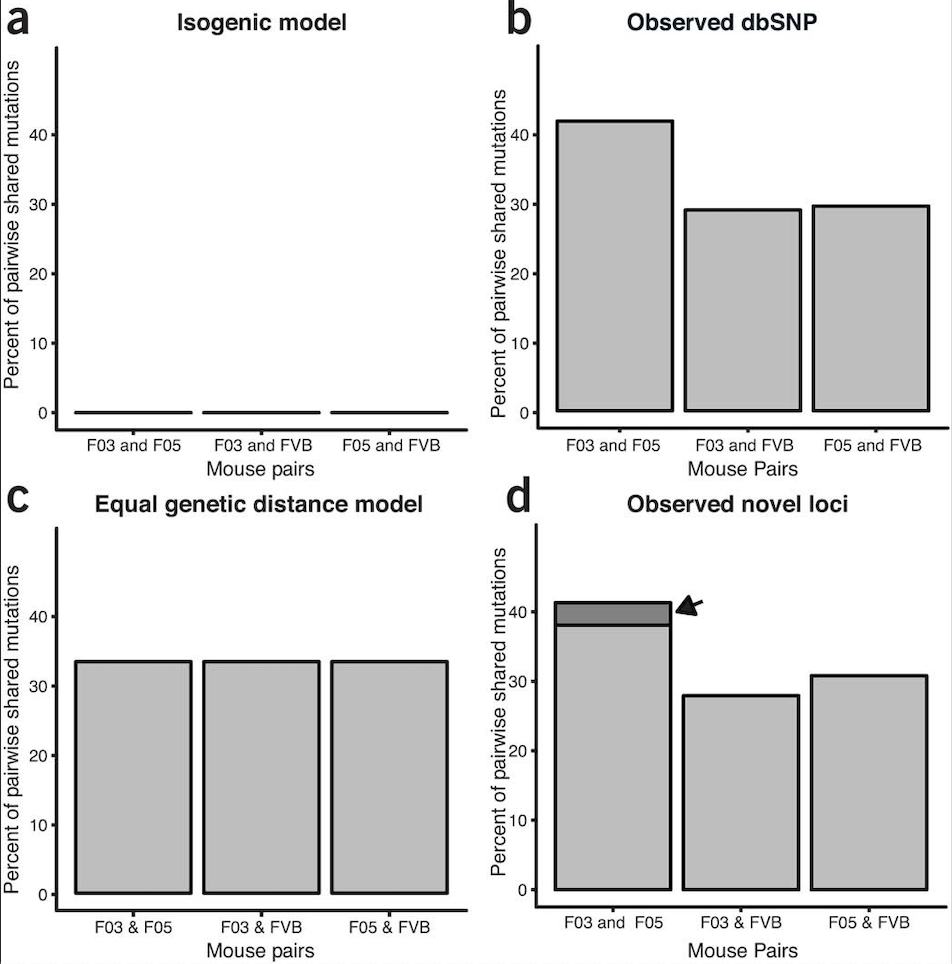
Response to “Unexpected mutations after CRISPR–Cas9 editing in vivo”
Nature Methods
·
30 Mar 2018
·
doi:10.1038/nmeth.4541
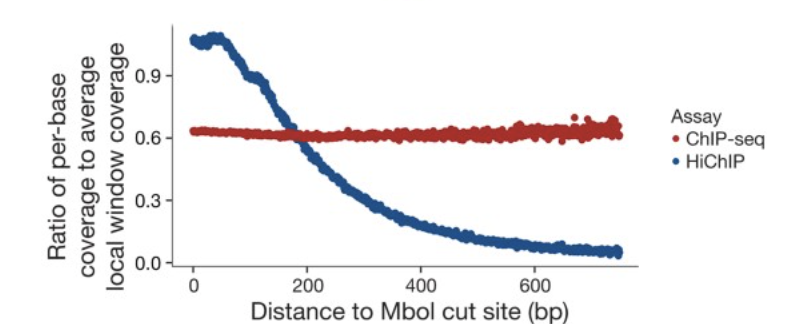
hichipper: a preprocessing pipeline for calling DNA loops from HiChIP data
Nature Methods
·
28 Feb 2018
·
doi:10.1038/nmeth.4583
2017
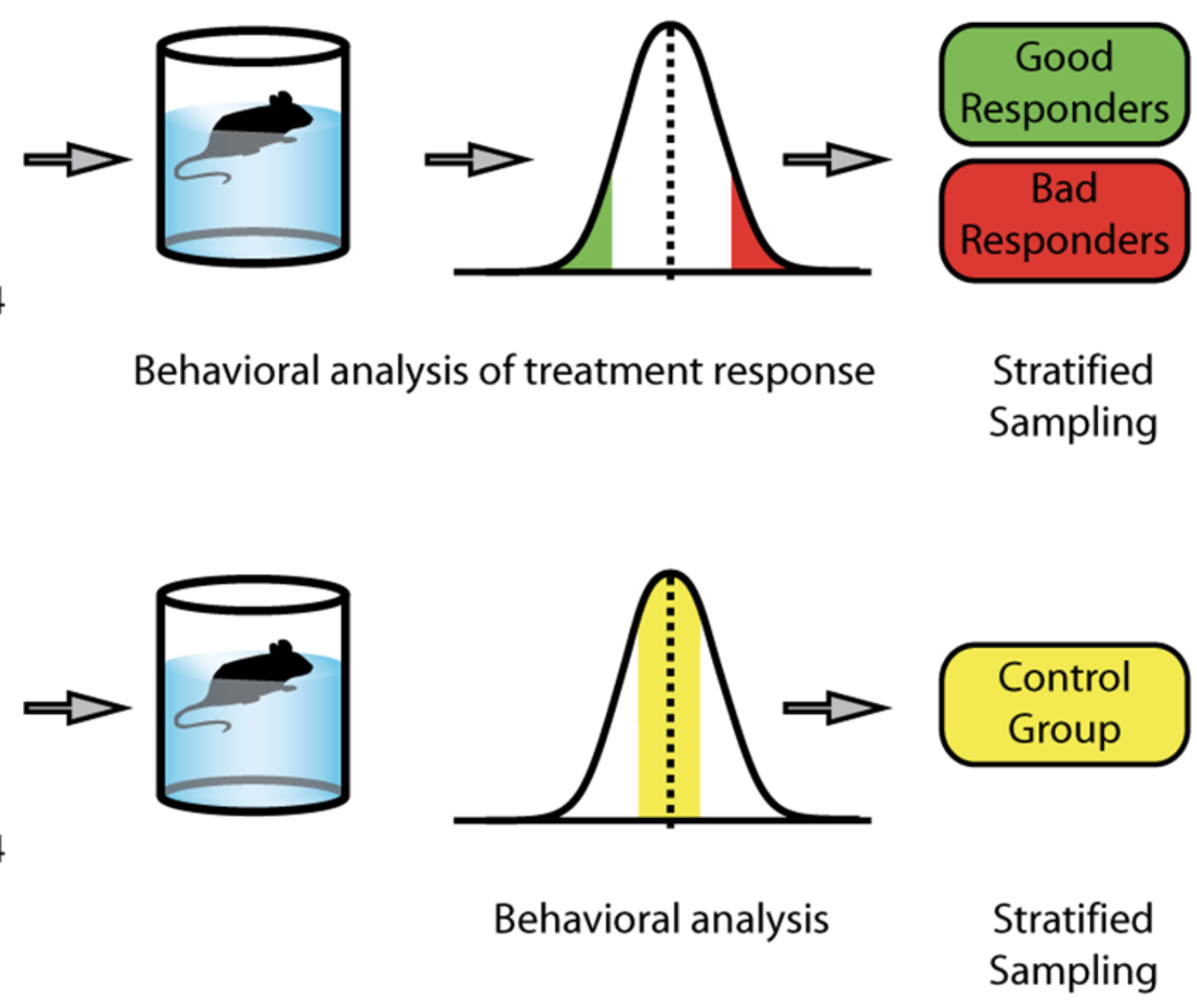
Common genes associated with antidepressant response in mouse and man identify key role of glucocorticoid receptor sensitivity
PLOS Biology
·
28 Dec 2017
·
doi:10.1371/journal.pbio.2002690
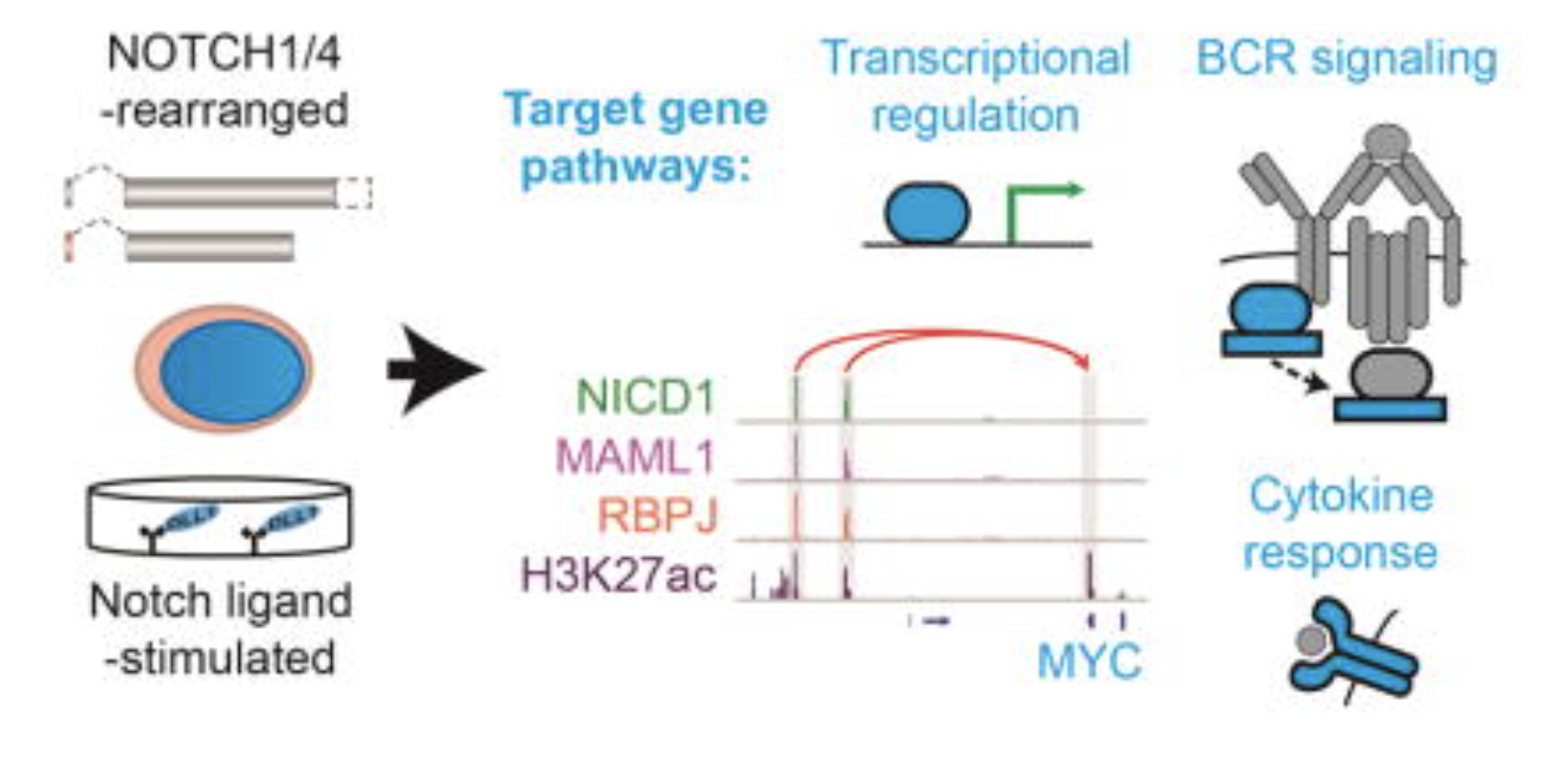
A B Cell Regulome Links Notch to Downstream Oncogenic Pathways in Small B Cell Lymphomas
Cell Reports
·
01 Oct 2017
·
doi:10.1016/j.celrep.2017.09.066
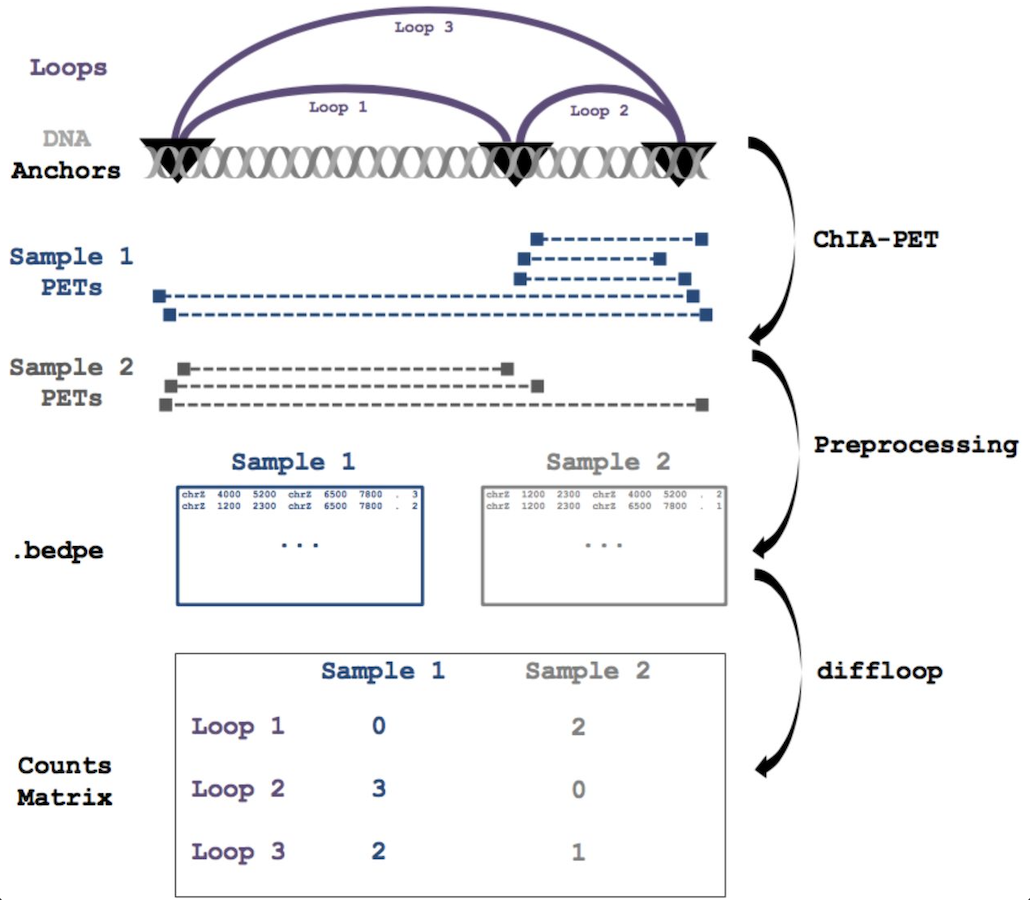
diffloop: a computational framework for identifying and analyzing differential DNA loops from sequencing data
Bioinformatics
·
29 Sep 2017
·
doi:10.1093/bioinformatics/btx623
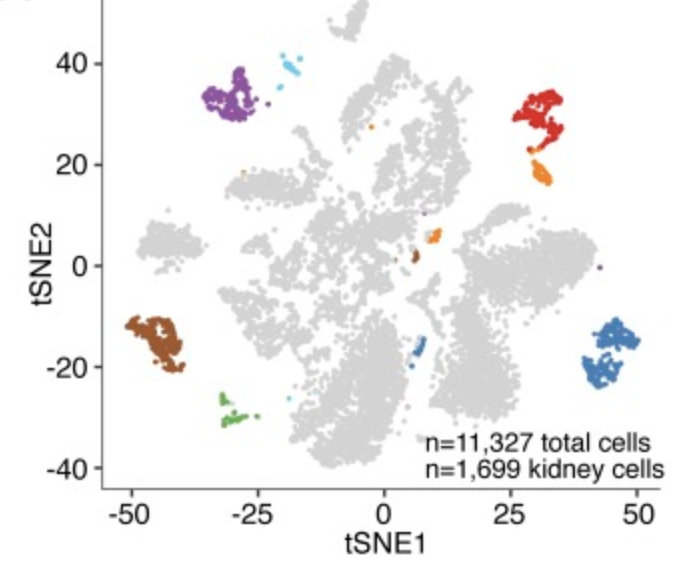
Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single-cell resolution using RNA sequencing
Journal of Experimental Medicine
·
06 Sep 2017
·
doi:10.1084/jem.20170976
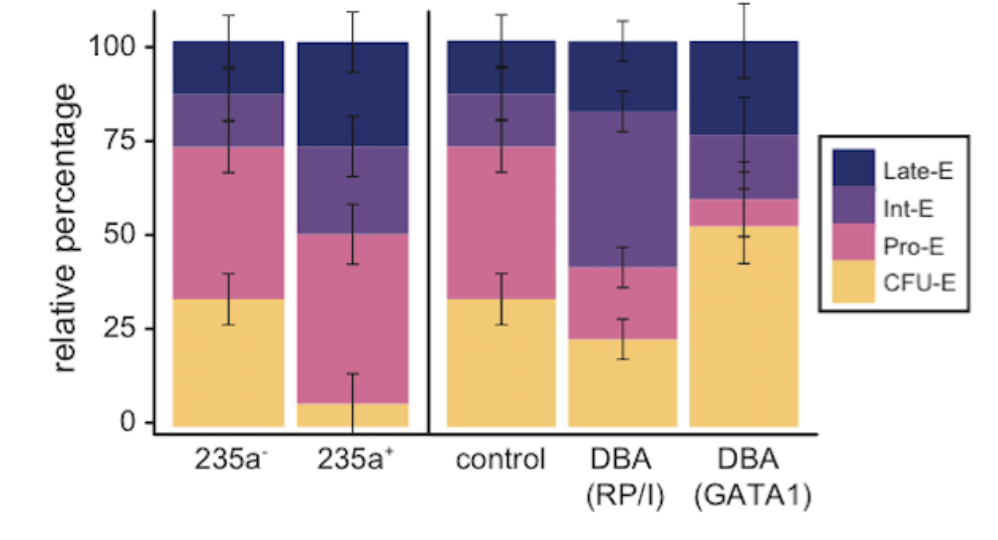
Confounding in ex vivo models of Diamond-Blackfan anemia
Blood
·
31 Aug 2017
·
doi:10.1182/blood-2017-05-783191
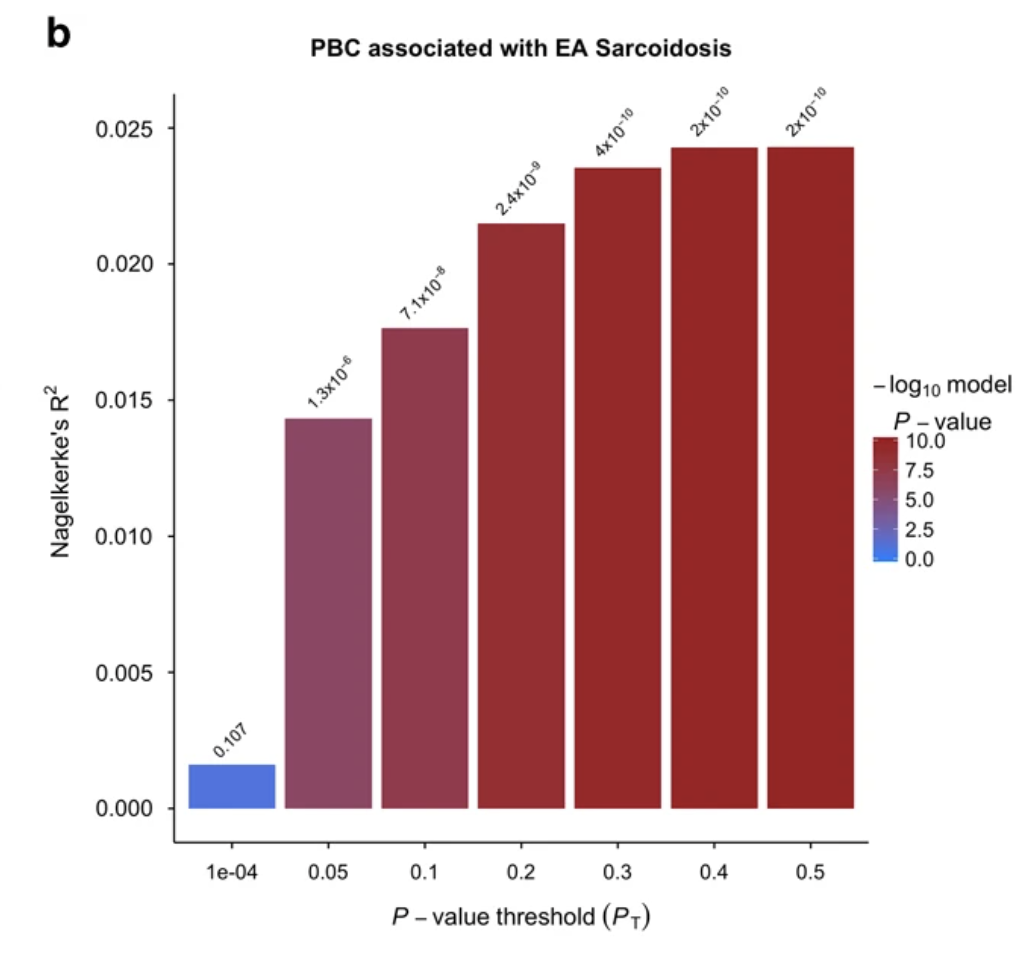
Polygenic risk assessment reveals pleiotropy between sarcoidosis and inflammatory disorders in the context of genetic ancestry
Genes & Immunity
·
01 Mar 2017
·
doi:10.1038/gene.2017.3
2016
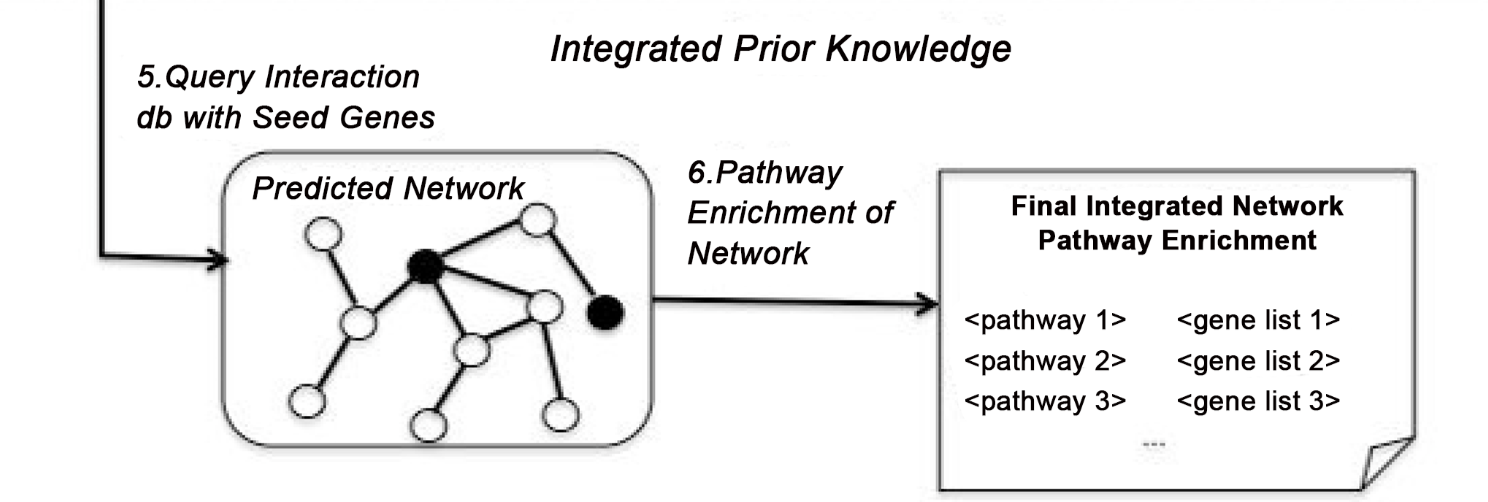
The Integration of Epistasis Network and Functional Interactions in a GWAS Implicates RXR Pathway Genes in the Immune Response to Smallpox Vaccine
PLOS ONE
·
11 Aug 2016
·
doi:10.1371/journal.pone.0158016
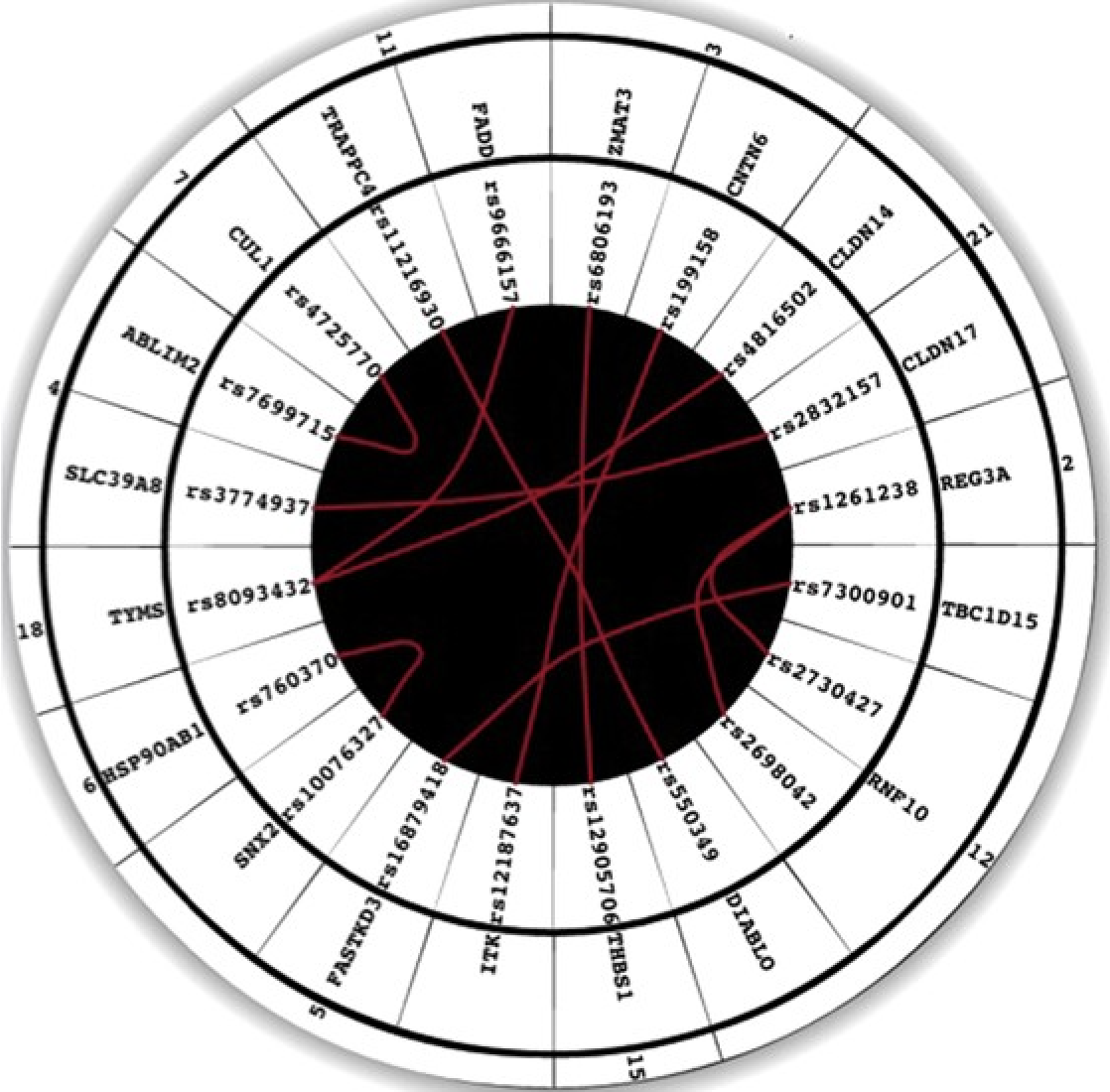
An interaction quantitative trait loci tool implicates epistatic functional variants in an apoptosis pathway in smallpox vaccine eQTL data
Genes & Immunity
·
07 Apr 2016
·
doi:10.1038/gene.2016.15
2015
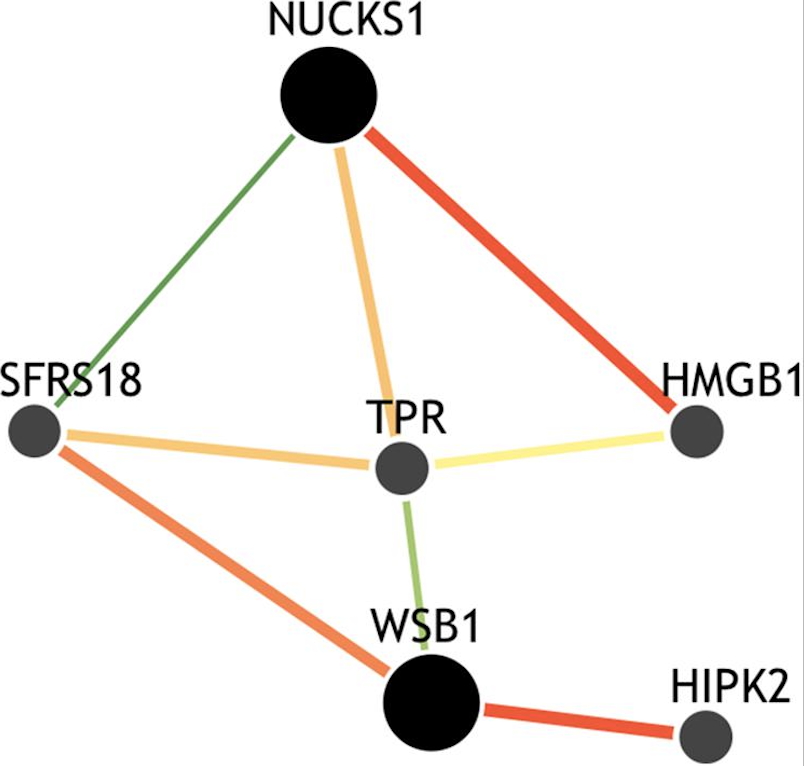
dcVar: a method for identifying common variants that modulate differential correlation structures in gene expression data
Frontiers in Genetics
·
19 Oct 2015
·
doi:10.3389/fgene.2015.00312
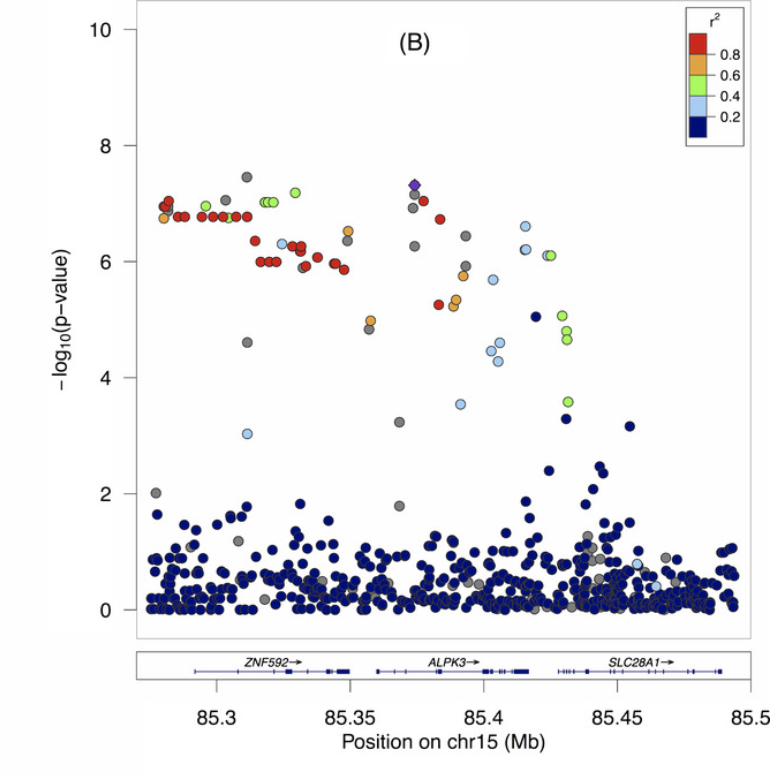
Fine mapping of chromosome 15q25 implicates ZNF 592 in neurosarcoidosis patients
Annals of Clinical and Translational Neurology
·
31 Jul 2015
·
doi:10.1002/acn3.229
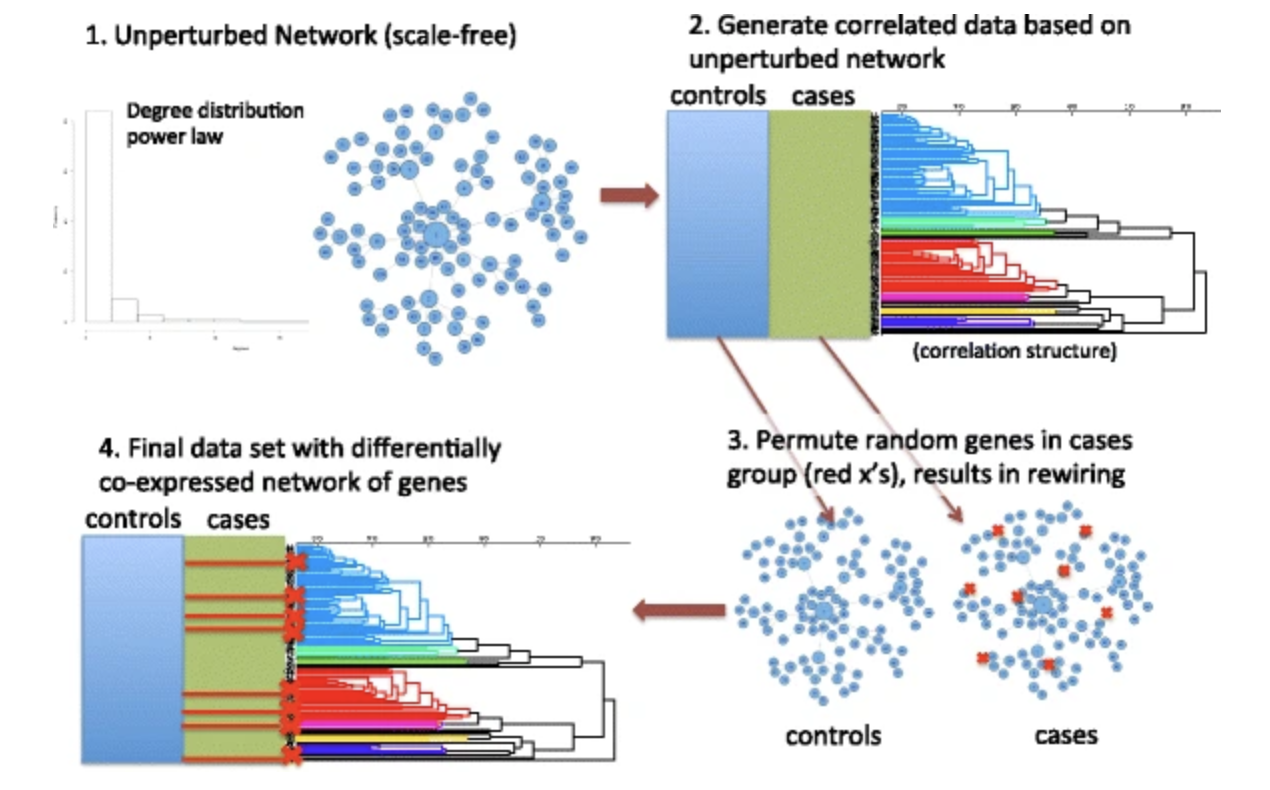
Differential co-expression network centrality and machine learning feature selection for identifying susceptibility hubs in networks with scale-free structure
BioData Mining
·
03 Feb 2015
·
doi:10.1186/s13040-015-0040-x
2014
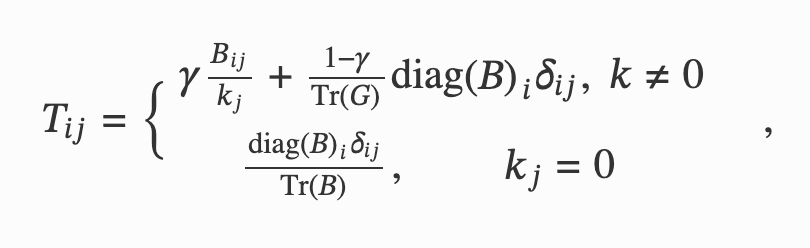
Network Theory for Data-Driven Epistasis Networks
Methods in Molecular Biology
·
03 Nov 2014
·
doi:10.1007/978-1-4939-2155-3_15
2013
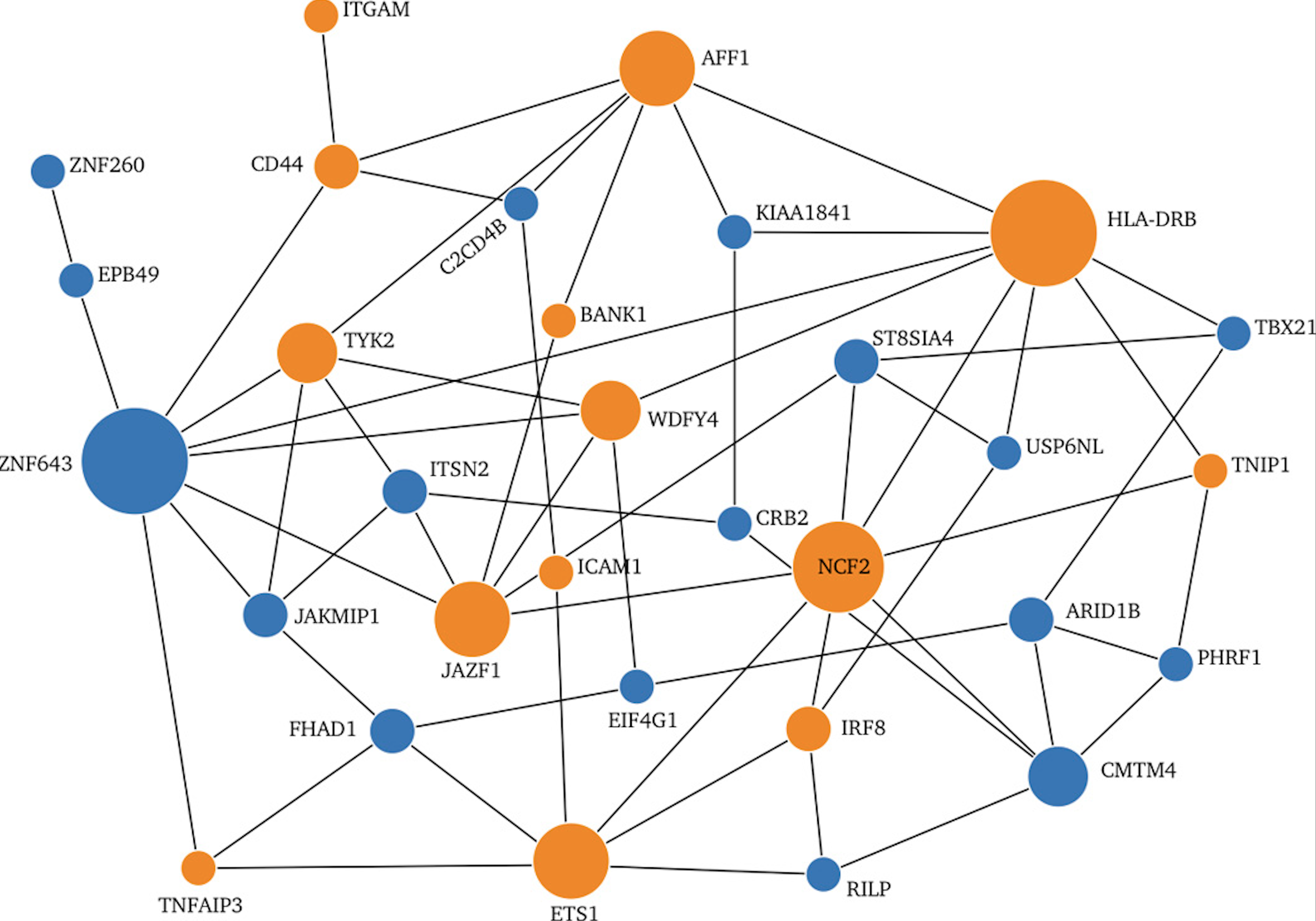
Encore: Genetic Association Interaction Network Centrality Pipeline and Application to SLE Exome Data
Genetic Epidemiology
·
05 Jun 2013
·
doi:10.1002/gepi.21739